|
|
|
|
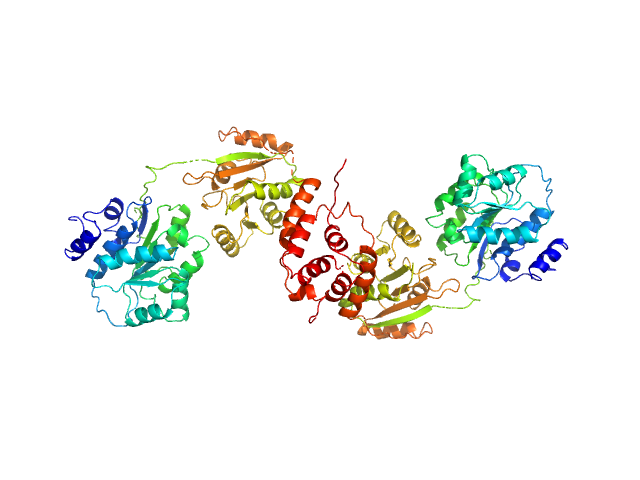
|
| Sample: |
Nucleolar RNA helicase 2 fragment 186-620 dimer, 98 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 500 mM NaCl, 10 % Glycerol, 2 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Mar 4
|
The Human RNA Helicase DDX21 Presents a Dimerization Interface Necessary for Helicase Activity
iScience 23(12):101811 (2020)
Marcaida M, Kauzlaric A, Duperrex A, Sülzle J, Moncrieffe M, Adebajo D, Manley S, Trono D, Dal Peraro M
|
| RgGuinier |
4.4 |
nm |
| Dmax |
16.9 |
nm |
| VolumePorod |
150 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleolar RNA helicase 2 monomer, 69 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 500 mM NaCl, 10 % Glycerol, 2 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Nov 3
|
The Human RNA Helicase DDX21 Presents a Dimerization Interface Necessary for Helicase Activity
iScience 23(12):101811 (2020)
Marcaida M, Kauzlaric A, Duperrex A, Sülzle J, Moncrieffe M, Adebajo D, Manley S, Trono D, Dal Peraro M
|
| RgGuinier |
4.1 |
nm |
| Dmax |
17.7 |
nm |
| VolumePorod |
111 |
nm3 |
|
|
|
|
|
|
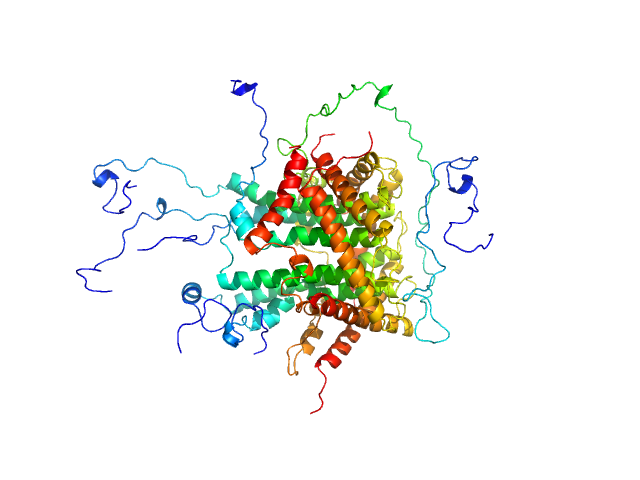
|
| Sample: |
Mitochondrial import inner membrane translocase subunit TIM8 trimer, 29 kDa Saccharomyces cerevisiae protein
Mitochondrial import inner membrane translocase subunit TIM13 trimer, 34 kDa Saccharomyces cerevisiae protein
Mitochondrial import inner membrane translocase subunit TIM23 monomer, 24 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Apr 24
|
Structural basis of client specificity in mitochondrial membrane-protein chaperones.
Sci Adv 6(51) (2020)
Sučec I, Wang Y, Dakhlaoui O, Weinhäupl K, Jores T, Costa D, Hessel A, Brennich M, Rapaport D, Lindorff-Larsen K, Bersch B, Schanda P
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.3 |
nm |
|
|
|
|
|
|
|
| Sample: |
Nucleolar RNA helicase 2 fragment 186-710 dimer, 121 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 500 mM NaCl, 10 % Glycerol, 2 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Mar 4
|
The Human RNA Helicase DDX21 Presents a Dimerization Interface Necessary for Helicase Activity
iScience 23(12):101811 (2020)
Marcaida M, Kauzlaric A, Duperrex A, Sülzle J, Moncrieffe M, Adebajo D, Manley S, Trono D, Dal Peraro M
|
| RgGuinier |
4.6 |
nm |
| Dmax |
17.7 |
nm |
| VolumePorod |
238 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Full-length HERV-K Rec response element monomer, 141 kDa Homo sapiens RNA
|
| Buffer: |
10mM Tris-HCl, 100mM KCl, and 0.5mM MgCl₂, pH: 8
|
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Feb 10
|
Structural Mimicry Drives HIV-1 Rev-Mediated HERV-K Expression
Journal of Molecular Biology 432(24):166711 (2020)
O'Carroll I, Fan L, Kroupa T, McShane E, Theodore C, Yates E, Kondrup B, Ding J, Martin T, Rein A, Wang Y
|
| RgGuinier |
7.5 |
nm |
| Dmax |
28.2 |
nm |
| VolumePorod |
454 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Truncated HERV-K Rec response element monomer, 102 kDa Homo sapiens RNA
|
| Buffer: |
10mM Tris-HCl, 100mM KCl, and 0.5mM MgCl₂, pH: 8
|
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Jul 17
|
Structural Mimicry Drives HIV-1 Rev-Mediated HERV-K Expression
Journal of Molecular Biology 432(24):166711 (2020)
O'Carroll I, Fan L, Kroupa T, McShane E, Theodore C, Yates E, Kondrup B, Ding J, Martin T, Rein A, Wang Y
|
| RgGuinier |
6.1 |
nm |
| Dmax |
21.8 |
nm |
| VolumePorod |
322 |
nm3 |
|
|
|
|
|
|
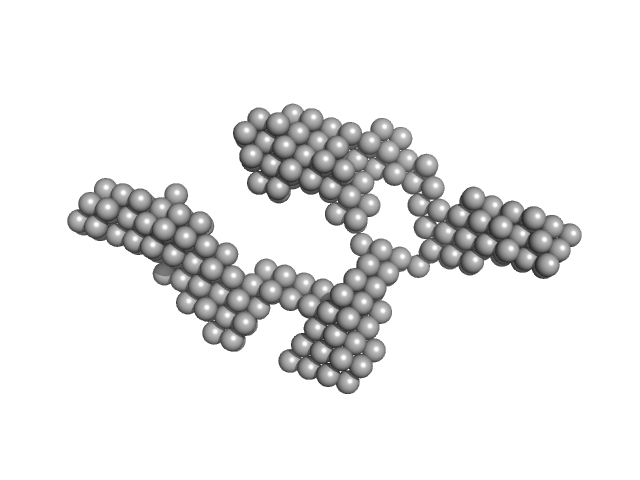
|
| Sample: |
TrΔSLIII HERV-K Rec response element monomer, 88 kDa Homo sapiens RNA
|
| Buffer: |
10mM Tris-HCl, 100mM KCl, and 0.5mM MgCl₂, pH: 8
|
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Dec 7
|
Structural Mimicry Drives HIV-1 Rev-Mediated HERV-K Expression
Journal of Molecular Biology 432(24):166711 (2020)
O'Carroll I, Fan L, Kroupa T, McShane E, Theodore C, Yates E, Kondrup B, Ding J, Martin T, Rein A, Wang Y
|
| RgGuinier |
5.3 |
nm |
| Dmax |
19.2 |
nm |
| VolumePorod |
226 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
TrΔSLIIab HERV-K Rec response element monomer, 86 kDa Homo sapiens RNA
|
| Buffer: |
10mM Tris-HCl, 100mM KCl, and 0.5mM MgCl₂, pH: 8
|
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Feb 21
|
Structural Mimicry Drives HIV-1 Rev-Mediated HERV-K Expression
Journal of Molecular Biology 432(24):166711 (2020)
O'Carroll I, Fan L, Kroupa T, McShane E, Theodore C, Yates E, Kondrup B, Ding J, Martin T, Rein A, Wang Y
|
| RgGuinier |
5.0 |
nm |
| Dmax |
18.4 |
nm |
| VolumePorod |
201 |
nm3 |
|
|
|
|
|
|
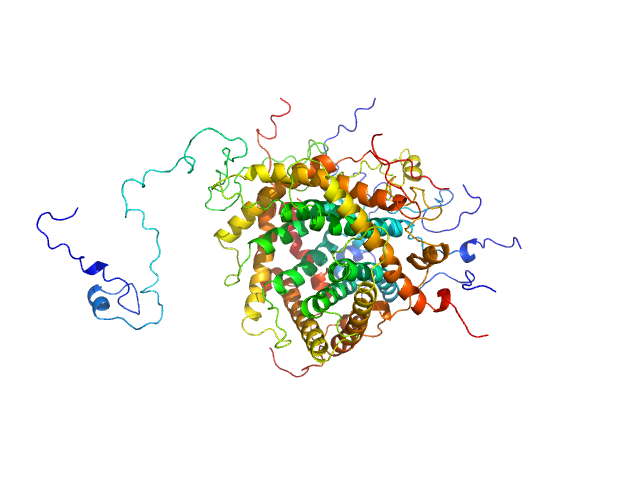
|
| Sample: |
Mitochondrial import inner membrane translocase subunit TIM9 trimer, 31 kDa Saccharomyces cerevisiae protein
Mitochondrial import inner membrane translocase subunit TIM10 trimer, 31 kDa Saccharomyces cerevisiae protein
Mitochondrial import inner membrane translocase subunit TIM23 monomer, 24 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 May 18
|
Structural basis of client specificity in mitochondrial membrane-protein chaperones.
Sci Adv 6(51) (2020)
Sučec I, Wang Y, Dakhlaoui O, Weinhäupl K, Jores T, Costa D, Hessel A, Brennich M, Rapaport D, Lindorff-Larsen K, Bersch B, Schanda P
|
| RgGuinier |
3.2 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
146 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Mitochondrial import inner membrane translocase subunit TIM8 trimer, 29 kDa Saccharomyces cerevisiae protein
Mitochondrial import inner membrane translocase subunit TIM13 trimer, 34 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Apr 21
|
Structural basis of client specificity in mitochondrial membrane-protein chaperones.
Sci Adv 6(51) (2020)
Sučec I, Wang Y, Dakhlaoui O, Weinhäupl K, Jores T, Costa D, Hessel A, Brennich M, Rapaport D, Lindorff-Larsen K, Bersch B, Schanda P
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.9 |
nm |
| VolumePorod |
136 |
nm3 |
|
|