|
|
|
|
|
| Sample: |
GraTA operator region monomer, 20 kDa DNA
antitoxin GraA, antidote of toxin GraT dimer, 22 kDa Pseudomonas putida protein
atitoxin GraA, antidote of GraT. dimer, 23 kDa Pseudomonas putida protein
|
| Buffer: |
50 mM Tris 250 mM NaCl 2 mM TCEP, pH: 8
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2017 Dec 17
|
A dual role in regulation and toxicity for the disordered N-terminus of the toxin GraT.
Nat Commun 10(1):972 (2019)
Talavera A, Tamman H, Ainelo A, Konijnenberg A, Hadži S, Sobott F, Garcia-Pino A, Hõrak R, Loris R
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
94 |
nm3 |
|
|
|
|
|
|
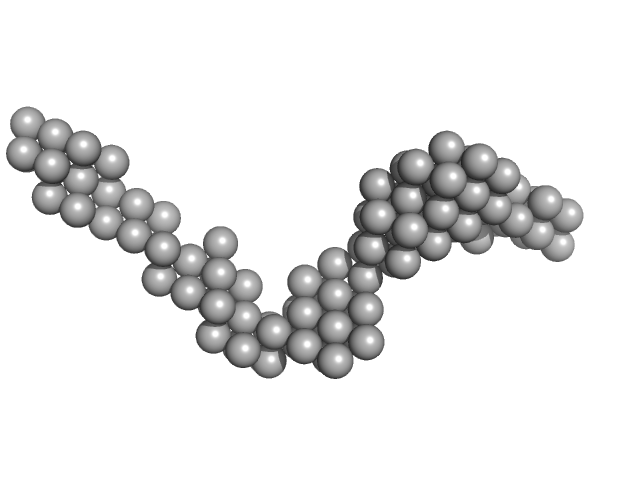
|
| Sample: |
octo-repeat PrP mRNA dimer, 144 kDa human PrP ORF RNA
|
| Buffer: |
10 mM Tris buffer, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Nov 4
|
Octa-repeat domain of the mammalian prion protein mRNA forms stable A-helical hairpin structure rather than G-quadruplexes.
Sci Rep 9(1):2465 (2019)
Czech A, Konarev PV, Goebel I, Svergun DI, Wills PR, Ignatova Z
|
| RgGuinier |
7.4 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
165 |
nm3 |
|
|
|
|
|
|
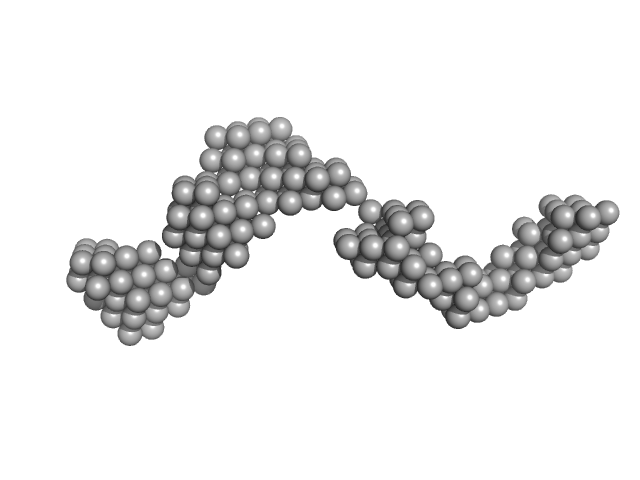
|
| Sample: |
octo-repeat PrP mRNA dimer, 144 kDa human PrP ORF RNA
|
| Buffer: |
10 mM Tris buffer with 100 mM KCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 6
|
Octa-repeat domain of the mammalian prion protein mRNA forms stable A-helical hairpin structure rather than G-quadruplexes.
Sci Rep 9(1):2465 (2019)
Czech A, Konarev PV, Goebel I, Svergun DI, Wills PR, Ignatova Z
|
| RgGuinier |
8.8 |
nm |
| Dmax |
31.0 |
nm |
| VolumePorod |
240 |
nm3 |
|
|
|
|
|
|
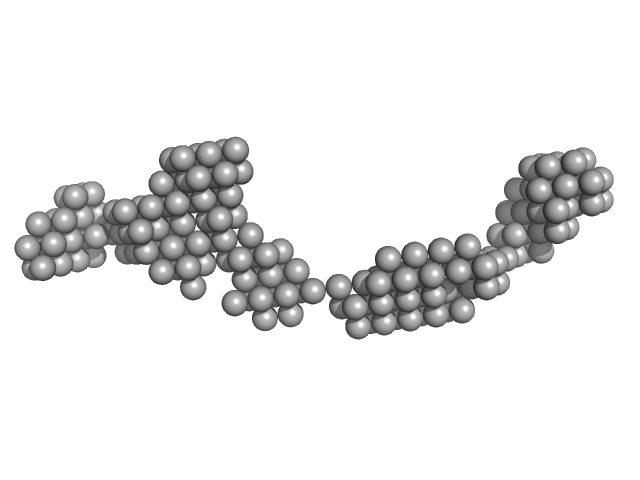
|
| Sample: |
octo-repeat PrP mRNA dimer, 144 kDa human PrP ORF RNA
|
| Buffer: |
10 mM Tris buffer with 100 mM LiCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 6
|
Octa-repeat domain of the mammalian prion protein mRNA forms stable A-helical hairpin structure rather than G-quadruplexes.
Sci Rep 9(1):2465 (2019)
Czech A, Konarev PV, Goebel I, Svergun DI, Wills PR, Ignatova Z
|
| RgGuinier |
9.0 |
nm |
| Dmax |
31.0 |
nm |
| VolumePorod |
225 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
octo-repeat PrP mRNA dimer, 144 kDa human PrP ORF RNA
|
| Buffer: |
10 mM Tris buffer with 100 mM KCl and 1 mM PDS, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 6
|
Octa-repeat domain of the mammalian prion protein mRNA forms stable A-helical hairpin structure rather than G-quadruplexes.
Sci Rep 9(1):2465 (2019)
Czech A, Konarev PV, Goebel I, Svergun DI, Wills PR, Ignatova Z
|
| RgGuinier |
9.1 |
nm |
| Dmax |
31.0 |
nm |
| VolumePorod |
220 |
nm3 |
|
|
|
|
|
|
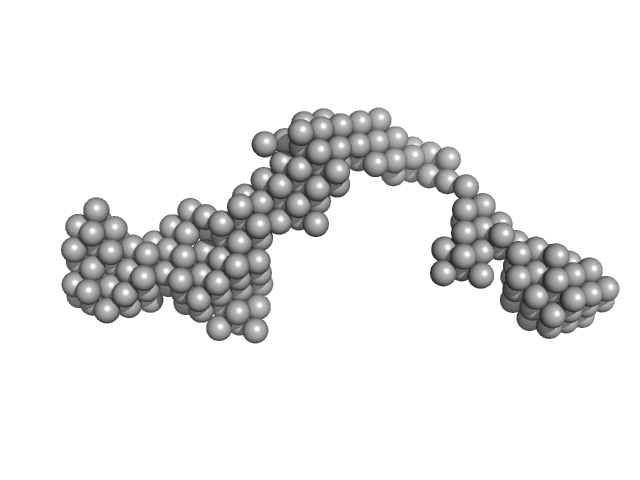
|
| Sample: |
octo-repeat PrP mRNA mutant dimer, 144 kDa human PrP ORF RNA
|
| Buffer: |
10 mM Tris buffer, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 6
|
Octa-repeat domain of the mammalian prion protein mRNA forms stable A-helical hairpin structure rather than G-quadruplexes.
Sci Rep 9(1):2465 (2019)
Czech A, Konarev PV, Goebel I, Svergun DI, Wills PR, Ignatova Z
|
| RgGuinier |
7.0 |
nm |
| Dmax |
24.0 |
nm |
| VolumePorod |
150 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
octo-repeat PrP mRNA mutant dimer, 144 kDa human PrP ORF RNA
|
| Buffer: |
10 mM Tris buffer with 100 mM KCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 6
|
Octa-repeat domain of the mammalian prion protein mRNA forms stable A-helical hairpin structure rather than G-quadruplexes.
Sci Rep 9(1):2465 (2019)
Czech A, Konarev PV, Goebel I, Svergun DI, Wills PR, Ignatova Z
|
| RgGuinier |
9.0 |
nm |
| Dmax |
33.0 |
nm |
| VolumePorod |
210 |
nm3 |
|
|
|
|
|
|
|
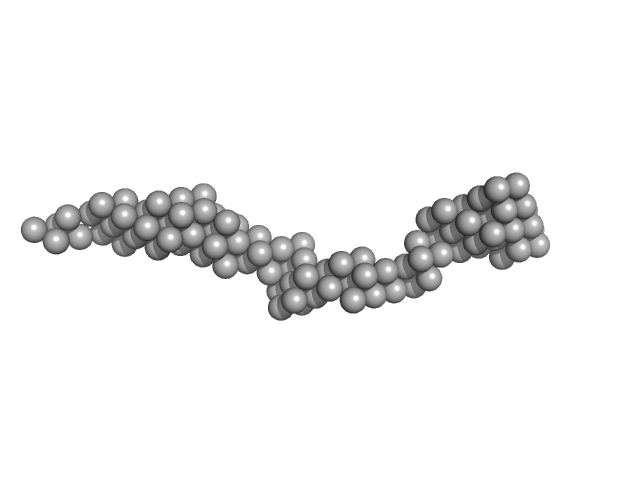
| Sample: |
octo-repeat PrP mRNA mutant dimer, 144 kDa human PrP ORF RNA
|
| Buffer: |
10 mM Tris buffer with 100 mM LiCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 6
|
Octa-repeat domain of the mammalian prion protein mRNA forms stable A-helical hairpin structure rather than G-quadruplexes.
Sci Rep 9(1):2465 (2019)
Czech A, Konarev PV, Goebel I, Svergun DI, Wills PR, Ignatova Z
|
| RgGuinier |
8.9 |
nm |
| Dmax |
33.0 |
nm |
| VolumePorod |
223 |
nm3 |
|
|
|
|
|
|
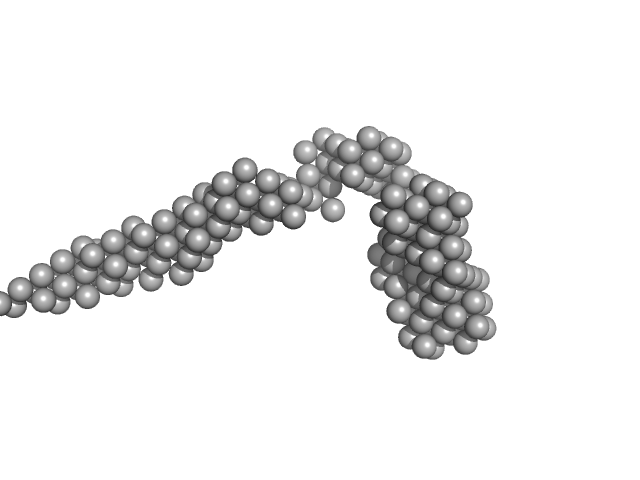
|
| Sample: |
octo-repeat PrP mRNA mutant dimer, 144 kDa human PrP ORF RNA
|
| Buffer: |
10 mM Tris buffer with 100 mM KCl and 1 mM PDS, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 6
|
Octa-repeat domain of the mammalian prion protein mRNA forms stable A-helical hairpin structure rather than G-quadruplexes.
Sci Rep 9(1):2465 (2019)
Czech A, Konarev PV, Goebel I, Svergun DI, Wills PR, Ignatova Z
|
| RgGuinier |
9.2 |
nm |
| Dmax |
33.0 |
nm |
| VolumePorod |
260 |
nm3 |
|
|
|
|
|
|
|
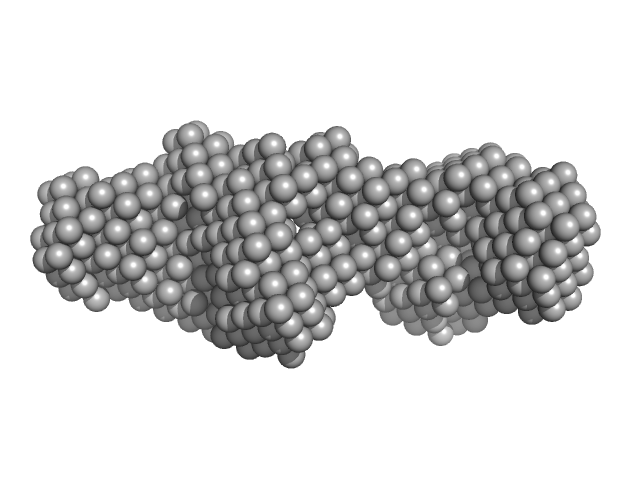
| Sample: |
LIM/homeobox protein Lhx4 monomer, 15 kDa Mus musculus protein
Insulin gene enhancer protein ISL-2 monomer, 4 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 8
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Nov 19
|
Mutation in a flexible linker modulates binding affinity for modular complexes.
Proteins (2019)
Stokes PH, Robertson NO, Silva AP, Estephan T, Trewhella J, Guss JM, Matthews JM
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
20 |
nm3 |
|
|