|
|
|
|
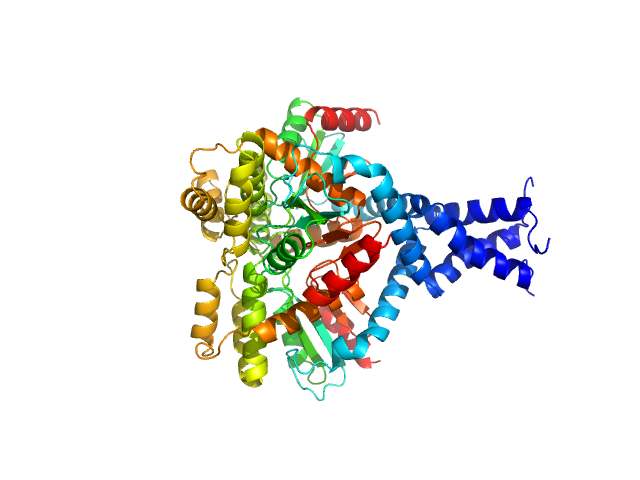
|
| Sample: |
Deletion mutant of PmScsC , 23 kDa Proteus mirabilis protein
|
| Buffer: |
10mM HEPES, 150mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Apr 5
|
Engineered variants provide new insight into the structural properties important for activity of the highly dynamic, trimeric protein disulfide isomerase ScsC from Proteus mirabilis.
Acta Crystallogr D Struct Biol 75(Pt 3):296-307 (2019)
Furlong EJ, Kurth F, Premkumar L, Whitten AE, Martin JL
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
|
|
|
|
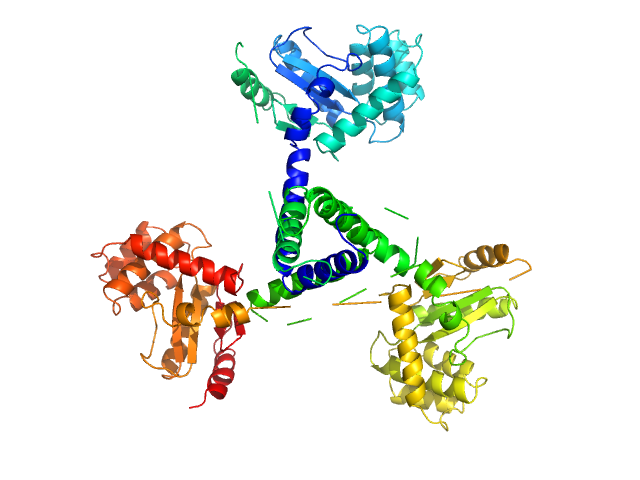
|
| Sample: |
Suppressor of Copper Sensitivity C protein trimer, 74 kDa Proteus mirabilis protein
|
| Buffer: |
10mM HEPES, 150mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Apr 5
|
Engineered variants provide new insight into the structural properties important for activity of the highly dynamic, trimeric protein disulfide isomerase ScsC from Proteus mirabilis.
Acta Crystallogr D Struct Biol 75(Pt 3):296-307 (2019)
Furlong EJ, Kurth F, Premkumar L, Whitten AE, Martin JL
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
101 |
nm3 |
|
|
|
|
|
|
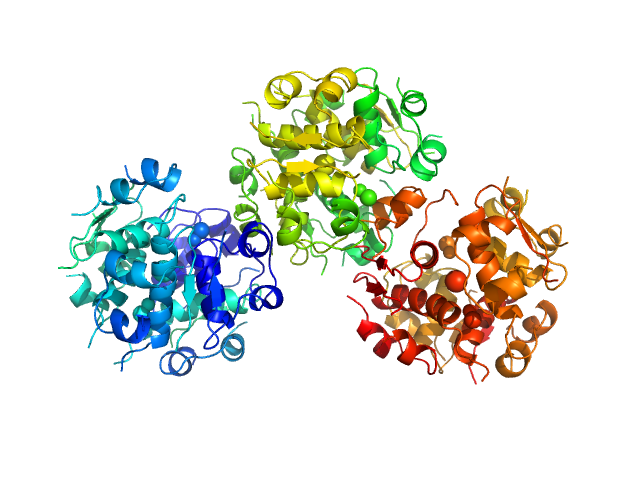
|
| Sample: |
Insulin detemir 18-mer, 106 kDa protein
|
| Buffer: |
5.0 mM Na2HPO4, 13.1 mM m-cresol, 15.1 mM phenol, 173.7 mM glycerol, 20.0 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at I911-4, MAX IV on 2015 Sep 25
|
Solution structures of long-acting insulin analogues and their complexes with albumin.
Acta Crystallogr D Struct Biol 75(Pt 3):272-282 (2019)
Ryberg LA, Sønderby P, Barrientos F, Bukrinski JT, Peters GHJ, Harris P
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.3 |
nm |
| VolumePorod |
137 |
nm3 |
|
|
|
|
|
|
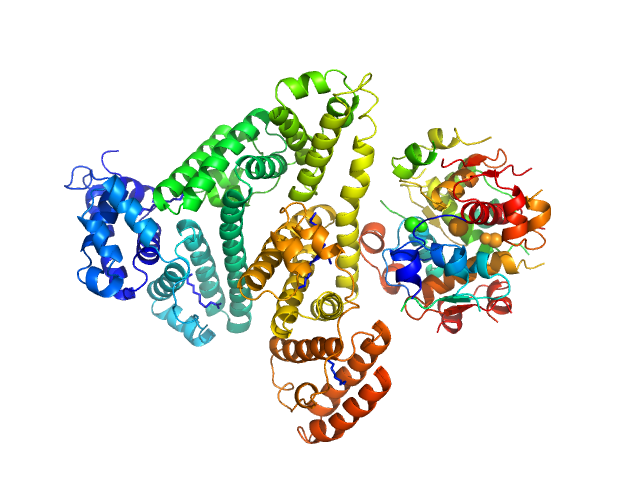
|
| Sample: |
Insulin detemir (Levemir(R), Novo Nordisk A/S) hexamer, 35 kDa protein
Human Albumin (Recombumin(R) Alpha, Albumedix Ltd.) monomer, 66 kDa protein
|
| Buffer: |
6.9 mM Na2HPO4, 11.9 mM m-cresol, 13.7 mM phenol, 157.3 mM glycerol, 38.5 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at I911-4, MAX IV on 2015 Sep 25
|
Solution structures of long-acting insulin analogues and their complexes with albumin.
Acta Crystallogr D Struct Biol 75(Pt 3):272-282 (2019)
Ryberg LA, Sønderby P, Barrientos F, Bukrinski JT, Peters GHJ, Harris P
|
| RgGuinier |
3.5 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
162 |
nm3 |
|
|
|
|
|
|
|
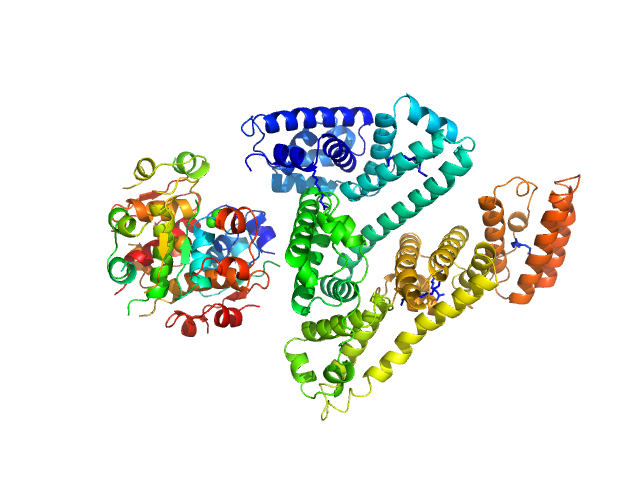
| Sample: |
Insulin detemir (Levemir(R), Novo Nordisk A/S) hexamer, 35 kDa protein
Human Albumin (Recombumin(R) Alpha, Albumedix Ltd.) monomer, 66 kDa protein
|
| Buffer: |
6.9 mM Na2HPO4, 11.9 mM m-cresol, 13.7 mM phenol, 157.3 mM glycerol, 38.5 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at I911-4, MAX IV on 2015 Sep 25
|
Solution structures of long-acting insulin analogues and their complexes with albumin.
Acta Crystallogr D Struct Biol 75(Pt 3):272-282 (2019)
Ryberg LA, Sønderby P, Barrientos F, Bukrinski JT, Peters GHJ, Harris P
|
| RgGuinier |
3.5 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
162 |
nm3 |
|
|
|
|
|
|
|
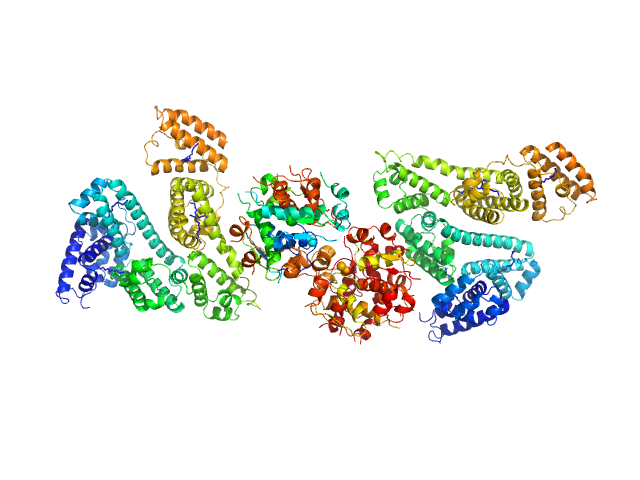
| Sample: |
Human Albumin (Recombumin(R) Alpha, Albumedix Ltd.) monomer, 66 kDa protein
Insulin detemir (Levemir(R), Novo Nordisk A/S) dodecamer, 71 kDa protein
|
| Buffer: |
8.8 mM Na2HPO4, 10.6 mM m-cresol, 12.2 mM phenol, 140.9 mM glycerol, 56.9 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at I911-4, MAX IV on 2015 Sep 25
|
Solution structures of long-acting insulin analogues and their complexes with albumin.
Acta Crystallogr D Struct Biol 75(Pt 3):272-282 (2019)
Ryberg LA, Sønderby P, Barrientos F, Bukrinski JT, Peters GHJ, Harris P
|
| RgGuinier |
5.4 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
309 |
nm3 |
|
|
|
|
|
|
|
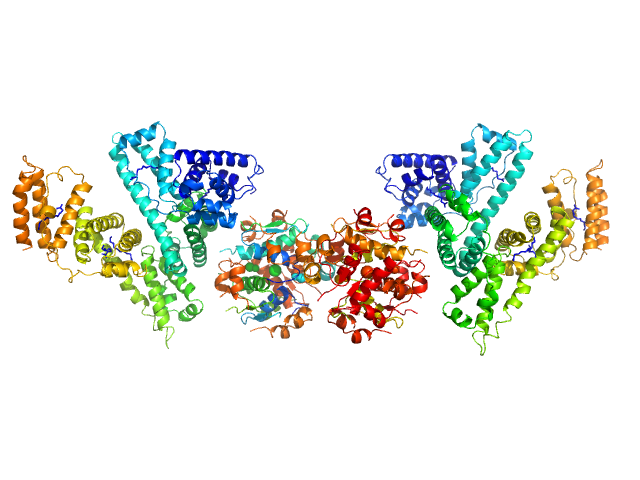
| Sample: |
Human Albumin (Recombumin(R) Alpha, Albumedix Ltd.) monomer, 66 kDa protein
Insulin detemir (Levemir(R), Novo Nordisk A/S) dodecamer, 71 kDa protein
|
| Buffer: |
8.8 mM Na2HPO4, 10.6 mM m-cresol, 12.2 mM phenol, 140.9 mM glycerol, 56.9 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at I911-4, MAX IV on 2015 Sep 25
|
Solution structures of long-acting insulin analogues and their complexes with albumin.
Acta Crystallogr D Struct Biol 75(Pt 3):272-282 (2019)
Ryberg LA, Sønderby P, Barrientos F, Bukrinski JT, Peters GHJ, Harris P
|
| RgGuinier |
5.4 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
309 |
nm3 |
|
|
|
|
|
|
|
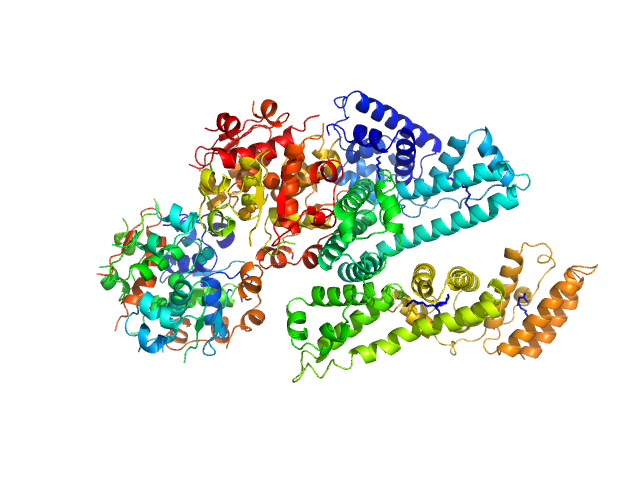
| Sample: |
Human Albumin (Recombumin(R) Elite, Albumedix Ltd.) monomer, 66 kDa protein
Insulin degludec(Tresiba(R), Novo Nordisk A/S) dodecamer, 73 kDa protein
|
| Buffer: |
25 mM Na2HPO4, 15.9 mM m-cresol, 15.9 mM phenol, 212.8 mM glycerol, 20 mM NaCl, pH: 7.6
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 27
|
Solution structures of long-acting insulin analogues and their complexes with albumin.
Acta Crystallogr D Struct Biol 75(Pt 3):272-282 (2019)
Ryberg LA, Sønderby P, Barrientos F, Bukrinski JT, Peters GHJ, Harris P
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.4 |
nm |
| VolumePorod |
179 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Putative killer protein, Toxin GraT. monomer, 11 kDa Pseudomonas putida protein
|
| Buffer: |
50 mM Tris 250 mM NaCl 2 mM TCEP, pH: 8
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2018 Sep 21
|
A dual role in regulation and toxicity for the disordered N-terminus of the toxin GraT.
Nat Commun 10(1):972 (2019)
Talavera A, Tamman H, Ainelo A, Konijnenberg A, Hadži S, Sobott F, Garcia-Pino A, Hõrak R, Loris R
|
| RgGuinier |
1.5 |
nm |
| Dmax |
5.5 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Putative killer protein, Toxin GraT. monomer, 11 kDa Pseudomonas putida protein
antitoxin GraA, antidote of toxin GraT dimer, 22 kDa Pseudomonas putida protein
Putative killer protein, Toxin GraT monomer, 11 kDa Pseudomonas putida protein
|
| Buffer: |
50 mM Tris 250 mM NaCl 2 mM TCEP, pH: 8
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jun 29
|
A dual role in regulation and toxicity for the disordered N-terminus of the toxin GraT.
Nat Commun 10(1):972 (2019)
Talavera A, Tamman H, Ainelo A, Konijnenberg A, Hadži S, Sobott F, Garcia-Pino A, Hõrak R, Loris R
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.9 |
nm |
| VolumePorod |
63 |
nm3 |
|
|