|
|
|
|
|
| Sample: |
Chromodomain-helicase-DNA-binding protein 1 dimer, 266 kDa Saccharomyces cerevisiae protein
169 bp DNA (145 bp Widom 601, flanked by 12bp DNA) monomer, 52 kDa DNA
Histone H2A type 1 monomer, 14 kDa Xenopus laevis protein
Histone H2B 1.1 monomer, 14 kDa Xenopus laevis protein
Histone H3.2 monomer, 15 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
|
| Buffer: |
10 mM Tris, 100 mM NaCl, 2 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 60% (w/v) sucrose, pH: 7.8
|
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 24
|
The ATPase motor of the Chd1 chromatin remodeler stimulates DNA unwrapping from the nucleosome.
Nucleic Acids Res 46(10):4978-4990 (2018)
Tokuda JM, Ren R, Levendosky RF, Tay RJ, Yan M, Pollack L, Bowman GD
|
| RgGuinier |
5.2 |
nm |
| Dmax |
12.8 |
nm |
|
|
|
|
|
|
|
| Sample: |
Chromodomain-helicase-DNA-binding protein 1 dimer, 266 kDa Saccharomyces cerevisiae protein
169 bp DNA (145 bp Widom 601, flanked by 12bp DNA) monomer, 52 kDa DNA
Histone H2A type 1 monomer, 14 kDa Xenopus laevis protein
Histone H2B 1.1 monomer, 14 kDa Xenopus laevis protein
Histone H3.2 monomer, 15 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
|
| Buffer: |
10 mM Tris, 100 mM NaCl, 2 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 60% (w/v) sucrose, ADP-BeF3 (0.5 mM ADP, 4 mM NaF, 0.6 mM BeCl2), pH: 7.8
|
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 24
|
The ATPase motor of the Chd1 chromatin remodeler stimulates DNA unwrapping from the nucleosome.
Nucleic Acids Res 46(10):4978-4990 (2018)
Tokuda JM, Ren R, Levendosky RF, Tay RJ, Yan M, Pollack L, Bowman GD
|
| RgGuinier |
5.3 |
nm |
| Dmax |
16.5 |
nm |
|
|
|
|
|
|
|
| Sample: |
Chromodomain-helicase-DNA-binding protein 1 dimer, 266 kDa Saccharomyces cerevisiae protein
169 bp DNA (145 bp Widom 601, flanked by 12bp DNA) monomer, 52 kDa DNA
Histone H2A type 1 monomer, 14 kDa Xenopus laevis protein
Histone H2B 1.1 monomer, 14 kDa Xenopus laevis protein
Histone H3.2 monomer, 15 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
|
| Buffer: |
10 mM Tris, 100 mM NaCl, 2 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 60% (w/v) sucrose, 0.5 mM AMP-PNP, pH: 7.8
|
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 24
|
The ATPase motor of the Chd1 chromatin remodeler stimulates DNA unwrapping from the nucleosome.
Nucleic Acids Res 46(10):4978-4990 (2018)
Tokuda JM, Ren R, Levendosky RF, Tay RJ, Yan M, Pollack L, Bowman GD
|
| RgGuinier |
5.6 |
nm |
| Dmax |
16.7 |
nm |
|
|
|
|
|
|

|
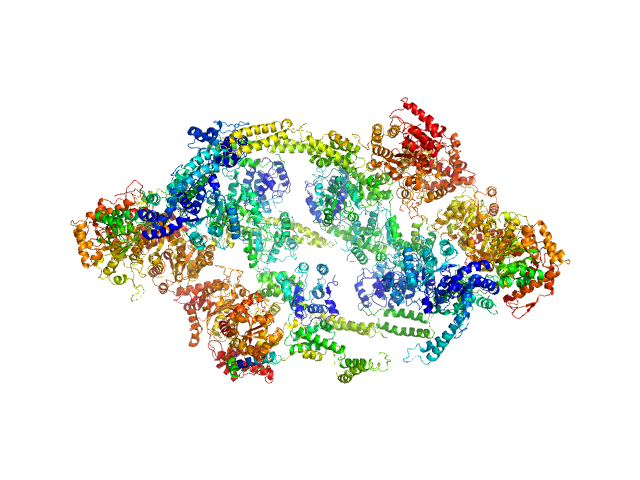
| Sample: |
Human telomerase RNA template component, DNA counterpart, nucleotides 1-20 dimer, 13 kDa Homo sapiens DNA
|
| Buffer: |
20 mM HEPES, 100 mM KCl,, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Aug 2
|
Structure and hydrodynamics of a DNA G-quadruplex with a cytosine bulge.
Nucleic Acids Res 46(10):5319-5331 (2018)
Meier M, Moya-Torres A, Krahn NJ, McDougall MD, Orriss GL, McRae EKS, Booy EP, McEleney K, Patel TR, McKenna SA, Stetefeld J
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.6 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
|
|
|
|
|
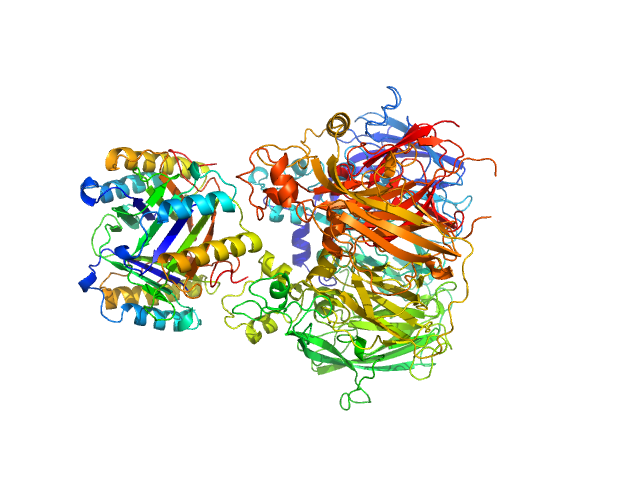
| Sample: |
ATP-dependent Clp protease ATP-binding subunit ClpC1 , 95 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
Hepes 50 mM pH 7.5, KCl 100 mM, glycerol 10%, MgCl2 4 mM and ATP 1 mM, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Sep 18
|
The antibiotic cyclomarin blocks arginine-phosphate-induced millisecond dynamics in the N-terminal domain of ClpC1 from Mycobacterium tuberculosis.
J Biol Chem 293(22):8379-8393 (2018)
Weinhäupl K, Brennich M, Kazmaier U, Lelievre J, Ballell L, Goldberg A, Schanda P, Fraga H
|
| RgGuinier |
7.6 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
2156 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
ATP-dependent Clp protease ATP-binding subunit ClpC1 , 95 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
Hepes 50 mM pH 7.5, KCl 100 mM, glycerol 10%, MgCl2 4 mM and ATP 1 mM, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Sep 18
|
The antibiotic cyclomarin blocks arginine-phosphate-induced millisecond dynamics in the N-terminal domain of ClpC1 from Mycobacterium tuberculosis.
J Biol Chem 293(22):8379-8393 (2018)
Weinhäupl K, Brennich M, Kazmaier U, Lelievre J, Ballell L, Goldberg A, Schanda P, Fraga H
|
| RgGuinier |
7.9 |
nm |
| Dmax |
25.1 |
nm |
| VolumePorod |
2416 |
nm3 |
|
|
|
|
|
|
|
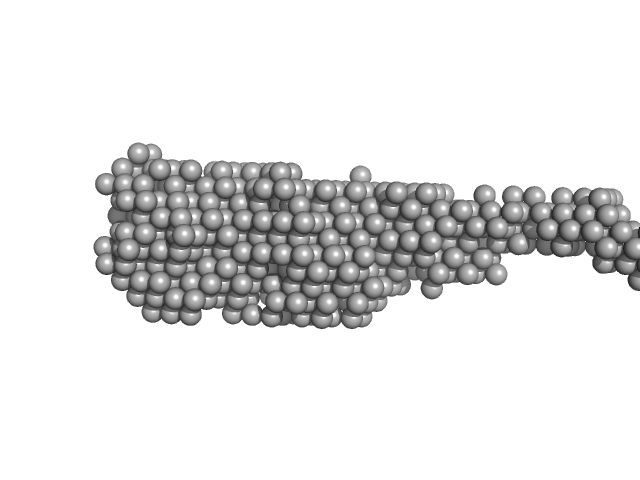
| Sample: |
Macrophage migration inhibitory factor trimer, 37 kDa Homo sapiens protein
Ceruloplasmin monomer, 122 kDa Homo sapiens protein
|
| Buffer: |
50mkm CuSO4, 100mM Hepes, pH: 7.4
|
| Experiment: |
SAXS
data collected at HECUS System-3, None on 2015 Dec 22
|
Structural Study of the Complex Formed by Ceruloplasmin and Macrophage Migration Inhibitory Factor.
Biochemistry (Mosc) 83(6):701-707 (2018)
Sokolov AV, Dadinova LA, Petoukhov MV, Bourenkov G, Dubova KM, Amarantov SV, Volkov VV, Kostevich VA, Gorbunov NP, Grudinina NA, Vasilyev VB, Samygina VR
|
| RgGuinier |
3.6 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
228 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
cGMP-dependent protein kinase 1 monomer, 70 kDa Bos taurus protein
|
| Buffer: |
50 mM MES, 300 mM NaCl, 1 mM TCEP, 5 mM DTT, pH: 6.9
|
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2015 Jun 6
|
An N-terminally truncated form of cyclic GMP-dependent protein kinase Iα (PKG Iα) is monomeric and autoinhibited and provides a model for activation.
J Biol Chem 293(21):7916-7929 (2018)
Moon TM, Sheehe JL, Nukareddy P, Nausch LW, Wohlfahrt J, Matthews DE, Blumenthal DK, Dostmann WR
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
105 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
e14A monomer, 21 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jun 16
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
2.8 |
nm |
| Dmax |
12.5 |
nm |
|
|
|
|
|
|

|
| Sample: |
e14B monomer, 13 kDa DNA
|
| Buffer: |
154 mM NaCl, pH: 8.3
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jun 16
|
Optical and Structural Characterization of a Chronic Myeloid Leukemia DNA Biosensor.
ACS Chem Biol 13(5):1235-1242 (2018)
Cordeiro M, Otrelo-Cardoso AR, Svergun DI, Konarev PV, Lima JC, Santos-Silva T, Baptista PV
|
| RgGuinier |
1.9 |
nm |
| Dmax |
9.0 |
nm |
|
|