|
|
|
|
|
| Sample: |
Leucine-rich repeat and fibronectin type-III domain-containing protein 4 dimer, 79 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris HCl, 100 mM NaCl, 0.02% NaN3,, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Jul 13
|
The structure of SALM5 suggests a dimeric assembly for the presynaptic RPTP ligand recognition.
Protein Eng Des Sel (2018)
Karki S, Paudel P, Sele C, Shkumatov AV, Kajander T
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
183 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Leucine-rich repeat and fibronectin type-III domain-containing protein 4 dimer, 109 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris HCl, 100 mM NaCl, 0.02% NaN3,, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Mar 11
|
The structure of SALM5 suggests a dimeric assembly for the presynaptic RPTP ligand recognition.
Protein Eng Des Sel (2018)
Karki S, Paudel P, Sele C, Shkumatov AV, Kajander T
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.1 |
nm |
| VolumePorod |
313 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Leucine-rich repeat and fibronectin type-III domain-containing protein 5 dimer, 82 kDa Mus musculus protein
|
| Buffer: |
30 mM Tris-Cl, 150 mM NaCl, 3% glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2016 Jun 8
|
The structure of SALM5 suggests a dimeric assembly for the presynaptic RPTP ligand recognition.
Protein Eng Des Sel (2018)
Karki S, Paudel P, Sele C, Shkumatov AV, Kajander T
|
| RgGuinier |
3.6 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
155 |
nm3 |
|
|
|
|
|
|

|
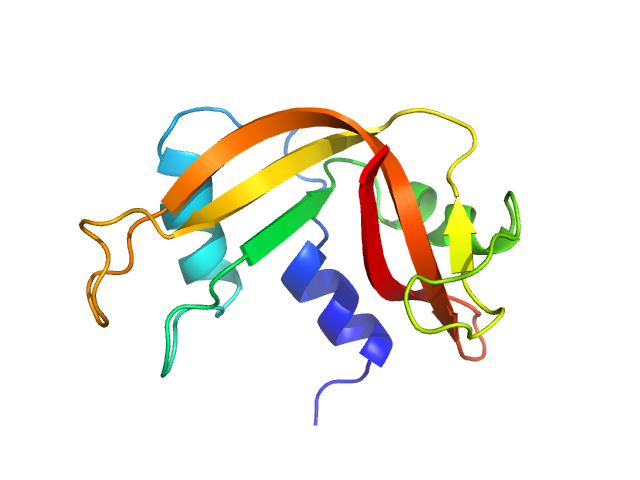
| Sample: |
Escherichia coli TraE protein (VirB8 homolog) hexamer, 171 kDa Escherichia coli protein
|
| Buffer: |
50 mM sodium phosphate 300 mM NaCl 40 mM imidazole 0.15 % octyl glucose neopentyl glycol (OGNG), pH: 7.4
|
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 Jun 2
|
VirB8 homolog TraE from plasmid pKM101 forms a hexameric ring structure and interacts with the VirB6 homolog TraD.
Proc Natl Acad Sci U S A 115(23):5950-5955 (2018)
Casu B, Mary C, Sverzhinsky A, Fouillen A, Nanci A, Baron C
|
| RgGuinier |
4.4 |
nm |
| Dmax |
13.7 |
nm |
| VolumePorod |
360 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ribonuclease pancreatic monomer, 16 kDa Bos taurus protein
|
| Buffer: |
phosphate buffered saline (PBS), pH: 7
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jul 29
|
Machine Learning Methods for X-Ray Scattering Data Analysis from Biomacromolecular Solutions.
Biophys J 114(11):2485-2492 (2018)
Franke D, Jeffries CM, Svergun DI
|
| RgGuinier |
1.6 |
nm |
| Dmax |
5.6 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Lipase B from Pseudozyma antarctica , 33 kDa Moesziomyces antarcticus protein
|
| Buffer: |
100 mM NaCl, 20 mM Na2HPO4, pH: 6
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jul 29
|
Machine Learning Methods for X-Ray Scattering Data Analysis from Biomacromolecular Solutions.
Biophys J 114(11):2485-2492 (2018)
Franke D, Jeffries CM, Svergun DI
|
|
|
|
|
|
|
|
| Sample: |
Lipase B from Pseudozyma antarctica , 33 kDa Moesziomyces antarcticus protein
|
| Buffer: |
100 mM NaCl, 20 mM Na2HPO4, 10 mM DTT, pH: 6
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jul 29
|
Machine Learning Methods for X-Ray Scattering Data Analysis from Biomacromolecular Solutions.
Biophys J 114(11):2485-2492 (2018)
Franke D, Jeffries CM, Svergun DI
|
|
|
|
|
|
|
|
| Sample: |
Ribonuclease pancreatic monomer, 16 kDa Bos taurus protein
|
| Buffer: |
10 mM HCl, pH: 1
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jul 29
|
Machine Learning Methods for X-Ray Scattering Data Analysis from Biomacromolecular Solutions.
Biophys J 114(11):2485-2492 (2018)
Franke D, Jeffries CM, Svergun DI
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Bovine serum albumin , 66 kDa Bos taurus protein
|
| Buffer: |
50 mM HEPES, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Sep 25
|
Machine Learning Methods for X-Ray Scattering Data Analysis from Biomacromolecular Solutions.
Biophys J 114(11):2485-2492 (2018)
Franke D, Jeffries CM, Svergun DI
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
169 bp DNA (145 bp Widom 601, flanked by 12bp DNA) monomer, 52 kDa DNA
Histone H2A type 1 monomer, 14 kDa Xenopus laevis protein
Histone H2B 1.1 monomer, 14 kDa Xenopus laevis protein
Histone H3.2 monomer, 15 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
|
| Buffer: |
10 mM Tris, 100 mM NaCl, 2 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 60% (w/v) sucrose, ADP-BeF3 (0.5 mM ADP, 4 mM NaF, 0.6 mM BeCl2), pH: 7.8
|
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 24
|
The ATPase motor of the Chd1 chromatin remodeler stimulates DNA unwrapping from the nucleosome.
Nucleic Acids Res 46(10):4978-4990 (2018)
Tokuda JM, Ren R, Levendosky RF, Tay RJ, Yan M, Pollack L, Bowman GD
|
| RgGuinier |
4.8 |
nm |
| Dmax |
14.0 |
nm |
|
|