|
|
|
|
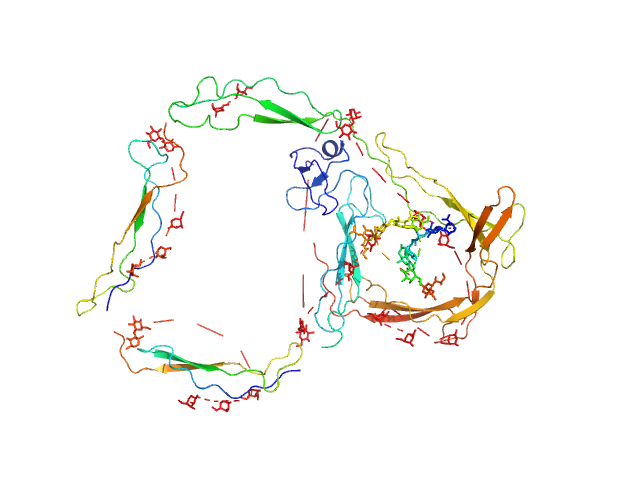
|
| Sample: |
E244K Human Properdin monomer, 49 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES 150 mM NaCl, pH: 7.2
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jul 27
|
Functional and structural insight into properdin control of complement alternative pathway amplification.
EMBO J 36(8):1084-1099 (2017)
Pedersen DV, Roumenina L, Jensen RK, Gadeberg TA, Marinozzi C, Picard C, Rybkine T, Thiel S, Sørensen UB, Stover C, Fremeaux-Bacchi V, Andersen GR
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
ESX-5 type VII secretion system protein EccC5 monomer, 142 kDa Mycobacterium xenopi RIVM700367 protein
|
| Buffer: |
20 mM Tris 200 mM NaCl 5%(v/v) Glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jul 15
|
Structure of the mycobacterial ESX-5 type VII secretion system membrane complex by single-particle analysis.
Nat Microbiol 2:17047 (2017)
Beckham KS, Ciccarelli L, Bunduc CM, Mertens HD, Ummels R, Lugmayr W, Mayr J, Rettel M, Savitski MM, Svergun DI, Bitter W, Wilmanns M, Marlovits TC, Parret AH, Houben EN
|
| RgGuinier |
6.2 |
nm |
| Dmax |
23.0 |
nm |
| VolumePorod |
255 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
GTPase ObgE/CgtA monomer, 44 kDa Escherichia coli protein
|
| Buffer: |
20 mM Hepes , 300 mM NaCl, 250 mM imidazole, 5 mM MgCl2, 2 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Jun 17
|
Structural and biochemical analysis of Escherichia coli ObgE, a central regulator of bacterial persistence.
J Biol Chem 292(14):5871-5883 (2017)
Gkekas S, Singh RK, Shkumatov AV, Messens J, Fauvart M, Verstraeten N, Michiels J, Versées W
|
| RgGuinier |
3.7 |
nm |
| Dmax |
18.1 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
GTPase ObgE/CgtA monomer, 44 kDa Escherichia coli protein
|
| Buffer: |
20 mM Hepes, 300 mM NaCl, 250 mM imidazole, 5 mM MgCl2, 2 mM DTT, 400 µM GppNHp, pH: 7.5
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Jun 17
|
Structural and biochemical analysis of Escherichia coli ObgE, a central regulator of bacterial persistence.
J Biol Chem 292(14):5871-5883 (2017)
Gkekas S, Singh RK, Shkumatov AV, Messens J, Fauvart M, Verstraeten N, Michiels J, Versées W
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
90 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
GTPase ObgE/CgtA monomer, 39 kDa Escherichia coli protein
|
| Buffer: |
20 mM Hepes , 300 mM NaCl, 250 mM imidazole, 5 mM MgCl2, 2 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Mar 12
|
Structural and biochemical analysis of Escherichia coli ObgE, a central regulator of bacterial persistence.
J Biol Chem 292(14):5871-5883 (2017)
Gkekas S, Singh RK, Shkumatov AV, Messens J, Fauvart M, Verstraeten N, Michiels J, Versées W
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
GTPase ObgE/CgtA monomer, 39 kDa Escherichia coli protein
|
| Buffer: |
20 mM Hepes, 300 mM NaCl, 250 mM imidazole, 5 mM MgCl2, 2 mM DTT, 400 µM GppNHp, pH: 7.5
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Mar 12
|
Structural and biochemical analysis of Escherichia coli ObgE, a central regulator of bacterial persistence.
J Biol Chem 292(14):5871-5883 (2017)
Gkekas S, Singh RK, Shkumatov AV, Messens J, Fauvart M, Verstraeten N, Michiels J, Versées W
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
72 |
nm3 |
|
|
|
|
|
|
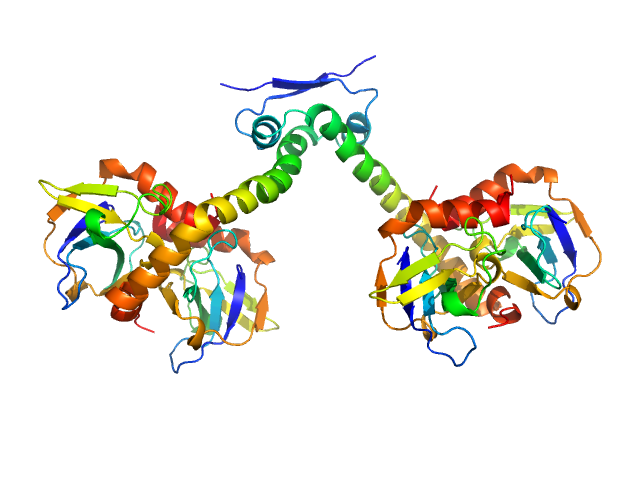
|
| Sample: |
Toxin CcdB dimer, 23 kDa Escherichia coli protein
Antitoxin CcdA dimer, 17 kDa Escherichia coli protein
Toxin CcdB dimer, 23 kDa Escherichia coli protein
|
| Buffer: |
10 mM Tris 50 mM NaCl, pH: 7.3
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2013 Jul 24
|
Molecular mechanism governing ratio-dependent transcription regulation in the ccdAB operon.
Nucleic Acids Res 45(6):2937-2950 (2017)
Vandervelde A, Drobnak I, Hadži S, Sterckx YG, Welte T, De Greve H, Charlier D, Efremov R, Loris R, Lah J
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
GTPase ObgE/CgtA monomer, 44 kDa Escherichia coli protein
|
| Buffer: |
20 mM Hepes, 300 mM NaCl, 250 mM imidazole, 5 mM MgCl2, 2 mM DTT, 400 uM GDP, pH: 7.5
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Jun 17
|
Structural and biochemical analysis of Escherichia coli ObgE, a central regulator of bacterial persistence.
J Biol Chem 292(14):5871-5883 (2017)
Gkekas S, Singh RK, Shkumatov AV, Messens J, Fauvart M, Verstraeten N, Michiels J, Versées W
|
| RgGuinier |
3.5 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
94673 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
GTPase ObgE/CgtA monomer, 39 kDa Escherichia coli protein
|
| Buffer: |
20 mM Hepes, 300 mM NaCl, 250 mM imidazole, 5 mM MgCl2, 2 mM DTT, 400 uM GDP, pH: 7.5
|
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, on 2015 Feb 1
|
Structural and biochemical analysis of Escherichia coli ObgE, a central regulator of bacterial persistence.
J Biol Chem 292(14):5871-5883 (2017)
Gkekas S, Singh RK, Shkumatov AV, Messens J, Fauvart M, Verstraeten N, Michiels J, Versées W
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
70673 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Type-2 restriction enzyme AgeI monomer, 31 kDa Thalassobius gelatinovorus protein
|
| Buffer: |
10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM CaCl₂, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 14
|
Restriction endonuclease AgeI is a monomer which dimerizes to cleave DNA.
Nucleic Acids Res 45(6):3547-3558 (2017)
Tamulaitiene G, Jovaisaite V, Tamulaitis G, Songailiene I, Manakova E, Zaremba M, Grazulis S, Xu SY, Siksnys V
|
| RgGuinier |
2.2 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
53 |
nm3 |
|
|