|
|
|
|
|
| Sample: |
Rab family protein dimer, 254 kDa Chlorobaculum tepidum protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl 5 mM MgCl2 5% Glycerol 1 mM DTT 200µM GppNHp, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Feb 16
|
A homologue of the Parkinson's disease-associated protein LRRK2 undergoes a monomer-dimer transition during GTP turnover.
Nat Commun 8(1):1008 (2017)
Deyaert E, Wauters L, Guaitoli G, Konijnenberg A, Leemans M, Terheyden S, Petrovic A, Gallardo R, Nederveen-Schippers LM, Athanasopoulos PS, Pots H, Van Haastert PJM, Sobott F, Gloeckner CJ, Efremov R...
|
| RgGuinier |
6.0 |
nm |
| Dmax |
23.0 |
nm |
| VolumePorod |
436 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Rab family protein dimer, 254 kDa Chlorobaculum tepidum protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl 5 mM MgCl2 5% Glycerol 1 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Feb 16
|
A homologue of the Parkinson's disease-associated protein LRRK2 undergoes a monomer-dimer transition during GTP turnover.
Nat Commun 8(1):1008 (2017)
Deyaert E, Wauters L, Guaitoli G, Konijnenberg A, Leemans M, Terheyden S, Petrovic A, Gallardo R, Nederveen-Schippers LM, Athanasopoulos PS, Pots H, Van Haastert PJM, Sobott F, Gloeckner CJ, Efremov R...
|
| RgGuinier |
5.0 |
nm |
| Dmax |
18.4 |
nm |
| VolumePorod |
447 |
nm3 |
|
|
|
|
|
|
|
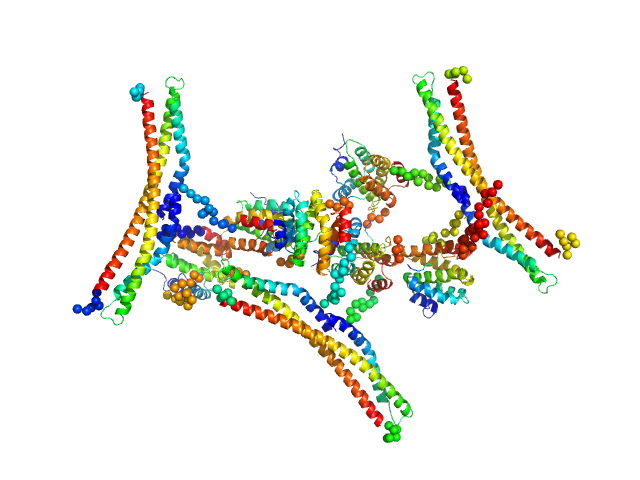
| Sample: |
Regulator of Ty1 transposition protein 103 dimer, 57 kDa Saccharomyces cerevisiae protein
DNA-directed RNA polymerase II subunit RPB1 monomer, 13 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
25 mM KH2PO4, 300 mM KCl, 10 mM BME, pH: 6.5
|
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2016 Apr 12
|
Structure and dynamics of the RNAPII CTDsome with Rtt103.
Proc Natl Acad Sci U S A 114(42):11133-11138 (2017)
Jasnovidova O, Klumpler T, Kubicek K, Kalynych S, Plevka P, Stefl R
|
| RgGuinier |
5.3 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
561 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Colicin N monomer, 43 kDa Escherichia coli protein
|
| Buffer: |
50 mM Na-Phosphate 300 mM NaCl, pH: 7.6
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2012 Jun 29
|
The Two-State Prehensile Tail of the Antibacterial Toxin Colicin N.
Biophys J 113(8):1673-1684 (2017)
Johnson CL, Solovyova AS, Hecht O, Macdonald C, Waller H, Grossmann JG, Moore GR, Lakey JH
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Colicin N delta 1-39 monomer, 39 kDa Escherichia coli protein
|
| Buffer: |
50 mM Na-Phosphate 300 mM NaCl, pH: 7.6
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2012 Jun 29
|
The Two-State Prehensile Tail of the Antibacterial Toxin Colicin N.
Biophys J 113(8):1673-1684 (2017)
Johnson CL, Solovyova AS, Hecht O, Macdonald C, Waller H, Grossmann JG, Moore GR, Lakey JH
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Colicin N K145A mutant monomer, 43 kDa Escherichia coli protein
|
| Buffer: |
50 mM Na-Phosphate 300 mM NaCl, pH: 7.6
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2012 Jun 29
|
The Two-State Prehensile Tail of the Antibacterial Toxin Colicin N.
Biophys J 113(8):1673-1684 (2017)
Johnson CL, Solovyova AS, Hecht O, Macdonald C, Waller H, Grossmann JG, Moore GR, Lakey JH
|
| RgGuinier |
3.2 |
nm |
| Dmax |
13.6 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Colicin N Translocation domain monomer, 10 kDa Escherichia coli protein
|
| Buffer: |
50 mM Na-Phosphate 300 mM NaCl, pH: 7.6
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2012 Jun 29
|
The Two-State Prehensile Tail of the Antibacterial Toxin Colicin N.
Biophys J 113(8):1673-1684 (2017)
Johnson CL, Solovyova AS, Hecht O, Macdonald C, Waller H, Grossmann JG, Moore GR, Lakey JH
|
| RgGuinier |
2.8 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
|
|
|
|
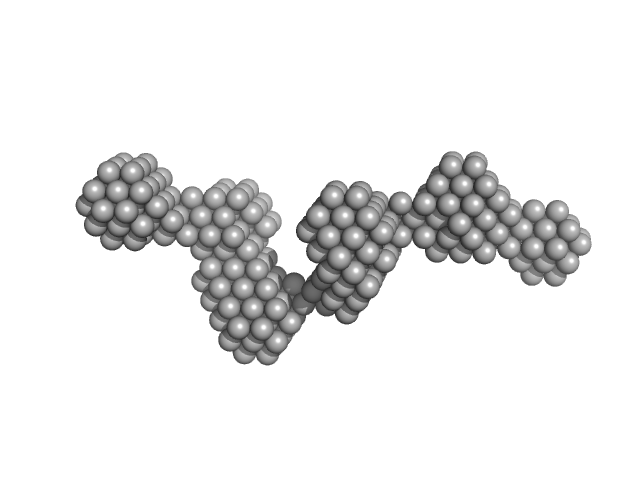
|
| Sample: |
Bacterial cellulose synthesis subunit C monomer, 71 kDa Enterobacter sp. CJF-002 protein
|
| Buffer: |
50 mM HEPES, 100 mM KCl, pH: 8
|
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2017 Apr 24
|
Crystal structure of the flexible tandem repeat domain of bacterial cellulose synthesis subunit C.
Sci Rep 7(1):13018 (2017)
Nojima S, Fujishima A, Kato K, Ohuchi K, Shimizu N, Yonezawa K, Tajima K, Yao M
|
| RgGuinier |
5.1 |
nm |
| Dmax |
18.5 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
|
|
|
|

|
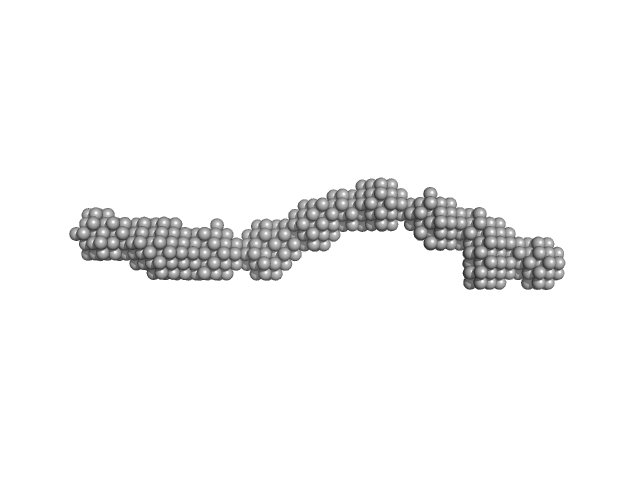
| Sample: |
CD22 extracellular domain monomer, 90 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl, pH: 9
|
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2016 Apr 21
|
Molecular basis of human CD22 function and therapeutic targeting.
Nat Commun 8(1):764 (2017)
Ereño-Orbea J, Sicard T, Cui H, Mazhab-Jafari MT, Benlekbir S, Guarné A, Rubinstein JL, Julien JP
|
| RgGuinier |
8.0 |
nm |
| Dmax |
30.6 |
nm |
|
|
|
|
|
|
|
| Sample: |
CD22 extracellular domain monomer, 90 kDa Homo sapiens protein
alpha(2,6)-Sialyllactose , 1 kDa
|
| Buffer: |
20 mM Tris 150 mM NaCl, pH: 9
|
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2016 Apr 21
|
Molecular basis of human CD22 function and therapeutic targeting.
Nat Commun 8(1):764 (2017)
Ereño-Orbea J, Sicard T, Cui H, Mazhab-Jafari MT, Benlekbir S, Guarné A, Rubinstein JL, Julien JP
|
| RgGuinier |
8.1 |
nm |
| Dmax |
29.8 |
nm |
|
|