|
|
|
|
|
| Sample: |
Gelsolin monomer, 84 kDa Homo sapiens protein
Actin, cytoplasmic 1 monomer, 42 kDa Gallus gallus protein
|
| Buffer: |
2 mM Tris-Cl, pH 8.0, 0.2 mM ATP, 1 mM NaN3, 0.1 mM CaCl2, 0.5 mM DTT, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Aug 31
|
Visualizing the nucleating and capped states of f-actin by Ca(2+)-gelsolin: Saxs data based structures of binary and ternary complexes.
Int J Biol Macromol :134556 (2024)
Sagar A, Peddada N, Choudhary V, Mir Y, Garg R, Ashish
|
| RgGuinier |
4.7 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
262 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Gelsolin monomer, 84 kDa Homo sapiens protein
Actin, cytoplasmic 1 monomer, 42 kDa Gallus gallus protein
|
| Buffer: |
2 mM Tris-Cl, pH 8.0, 0.2 mM ATP, 1 mM NaN3, 0.1 mM CaCl2, 0.5 mM DTT, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Aug 31
|
Visualizing the nucleating and capped states of f-actin by Ca(2+)-gelsolin: Saxs data based structures of binary and ternary complexes.
Int J Biol Macromol :134556 (2024)
Sagar A, Peddada N, Choudhary V, Mir Y, Garg R, Ashish
|
| RgGuinier |
5.2 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
266 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Gelsolin monomer, 84 kDa Homo sapiens protein
Actin, cytoplasmic 1 monomer, 42 kDa Gallus gallus protein
|
| Buffer: |
2 mM Tris-Cl, pH 8.0, 0.2 mM ATP, 1 mM NaN3, 0.1 mM CaCl2, 0.5 mM DTT, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Aug 31
|
Visualizing the nucleating and capped states of f-actin by Ca(2+)-gelsolin: Saxs data based structures of binary and ternary complexes.
Int J Biol Macromol :134556 (2024)
Sagar A, Peddada N, Choudhary V, Mir Y, Garg R, Ashish
|
| RgGuinier |
4.4 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
241 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Gelsolin monomer, 84 kDa Homo sapiens protein
Actin, cytoplasmic 1 monomer, 42 kDa Gallus gallus protein
|
| Buffer: |
2 mM Tris-Cl, pH 8.0, 0.2 mM ATP, 1 mM NaN3, 0.1 mM CaCl2, 0.5 mM DTT, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Sep 1
|
Visualizing the nucleating and capped states of f-actin by Ca(2+)-gelsolin: Saxs data based structures of binary and ternary complexes.
Int J Biol Macromol :134556 (2024)
Sagar A, Peddada N, Choudhary V, Mir Y, Garg R, Ashish
|
| RgGuinier |
4.6 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
243 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA repair protein RAD52 homolog undecamer, 528 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 7.5, 250 mM NaCl, 1% glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Jul 21
|
An integrative structural study of the human full-length RAD52 at 2.2 Å resolution
Communications Biology 7(1) (2024)
Balboni B, Marotta R, Rinaldi F, Milordini G, Varignani G, Girotto S, Cavalli A
|
| RgGuinier |
8.0 |
nm |
| Dmax |
40.2 |
nm |
| VolumePorod |
1218 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
1-deoxy-D-xylulose-5-phosphate reductoisomerase (G328C, K361E, S551G) dimer, 104 kDa Toxoplasma gondii (strain … protein
|
| Buffer: |
20 mM Tris/HCl, 150 mM NaCl, 40 mM MgCl2, 2% glycerol,, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Apr 25
|
1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR) as target for anti Toxoplasma gondii
agents: crystal structure, biochemical characterisation and biological evaluation of inhibitors
Biochemical Journal (2024)
Mazzone F, Hoeppner A, Reiners J, Gertzen C, Applegate V, Abdullaziz M, Gottstein J, Degrandi D, Wesemann M, Kurz T, Smits S, Pfeffer K
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.4 |
nm |
| VolumePorod |
176 |
nm3 |
|
|
|
|
|
|
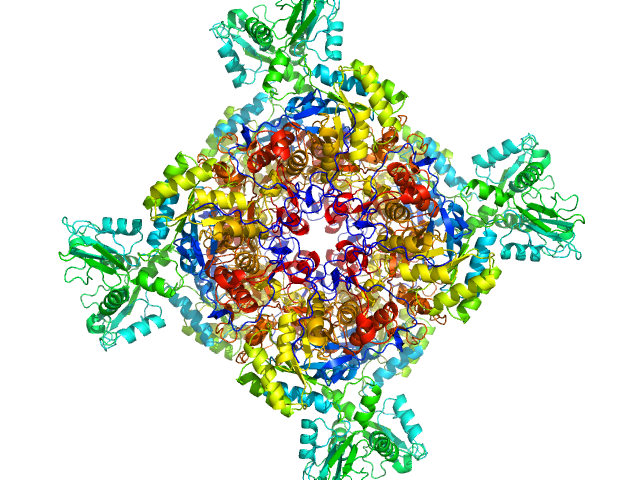
|
| Sample: |
Inosine-5'-monophosphate dehydrogenase octamer, 426 kDa Mycolicibacterium smegmatis (strain … protein
|
| Buffer: |
50 mM HEPES, 200 mM KCl, 2 mM MgCl2, 0.5 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2024 Feb 21
|
Deciphering the allosteric regulation of mycobacterial inosine-5′-monophosphate dehydrogenase
Nature Communications 15(1) (2024)
Bulvas O, Knejzlík Z, Sýs J, Filimoněnko A, Čížková M, Clarová K, Rejman D, Kouba T, Pichová I
|
| RgGuinier |
5.3 |
nm |
| Dmax |
24.5 |
nm |
| VolumePorod |
952 |
nm3 |
|
|
|
|
|
|
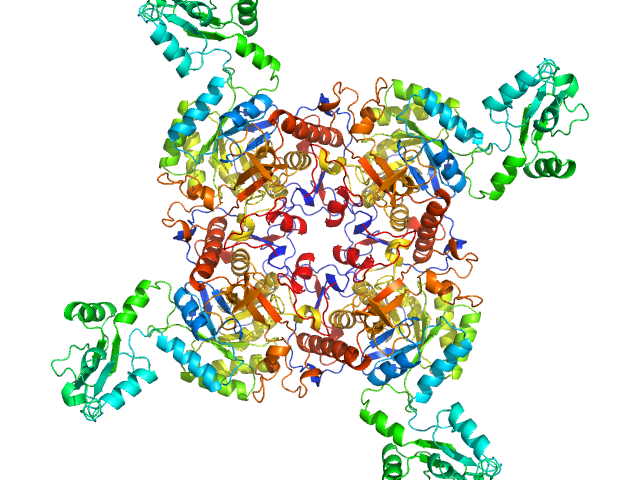
|
| Sample: |
Inosine-5'-monophosphate dehydrogenase tetramer, 213 kDa Mycolicibacterium smegmatis (strain … protein
|
| Buffer: |
50 mM HEPES, 200 mM KCl, 2 mM MgCl2, 0.5 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2024 Mar 6
|
Deciphering the allosteric regulation of mycobacterial inosine-5′-monophosphate dehydrogenase
Nature Communications 15(1) (2024)
Bulvas O, Knejzlík Z, Sýs J, Filimoněnko A, Čížková M, Clarová K, Rejman D, Kouba T, Pichová I
|
| RgGuinier |
5.0 |
nm |
| Dmax |
21.5 |
nm |
| VolumePorod |
471 |
nm3 |
|
|
|
|
|
|
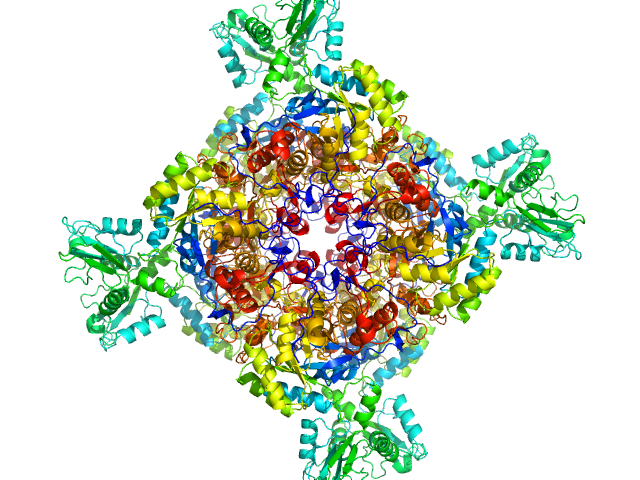
|
| Sample: |
Inosine-5'-monophosphate dehydrogenase octamer, 426 kDa Mycolicibacterium smegmatis (strain … protein
|
| Buffer: |
50 mM HEPES, 200 mM KCl, 2 mM MgCl2, 0.5 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2024 Jan 29
|
Deciphering the allosteric regulation of mycobacterial inosine-5′-monophosphate dehydrogenase
Nature Communications 15(1) (2024)
Bulvas O, Knejzlík Z, Sýs J, Filimoněnko A, Čížková M, Clarová K, Rejman D, Kouba T, Pichová I
|
| RgGuinier |
5.0 |
nm |
| Dmax |
14.8 |
nm |
| VolumePorod |
821 |
nm3 |
|
|
|
|
|
|
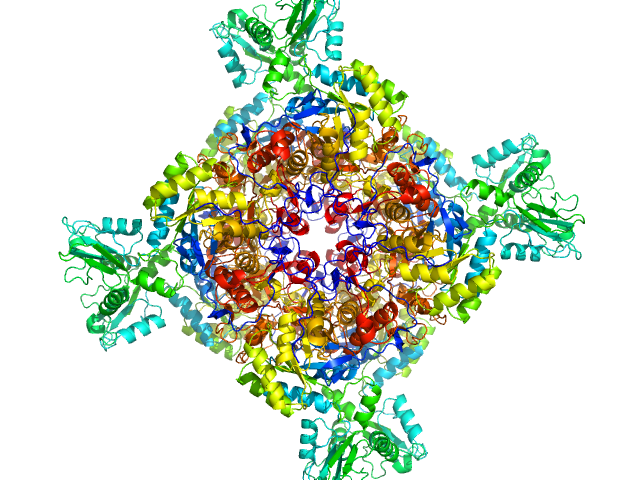
|
| Sample: |
Inosine-5'-monophosphate dehydrogenase octamer, 426 kDa Mycolicibacterium smegmatis (strain … protein
|
| Buffer: |
50 mM HEPES, 200 mM KCl, 2 mM MgCl2, 0.5 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2024 Mar 6
|
Deciphering the allosteric regulation of mycobacterial inosine-5′-monophosphate dehydrogenase
Nature Communications 15(1) (2024)
Bulvas O, Knejzlík Z, Sýs J, Filimoněnko A, Čížková M, Clarová K, Rejman D, Kouba T, Pichová I
|
| RgGuinier |
5.1 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
784 |
nm3 |
|
|