|
|
|
|
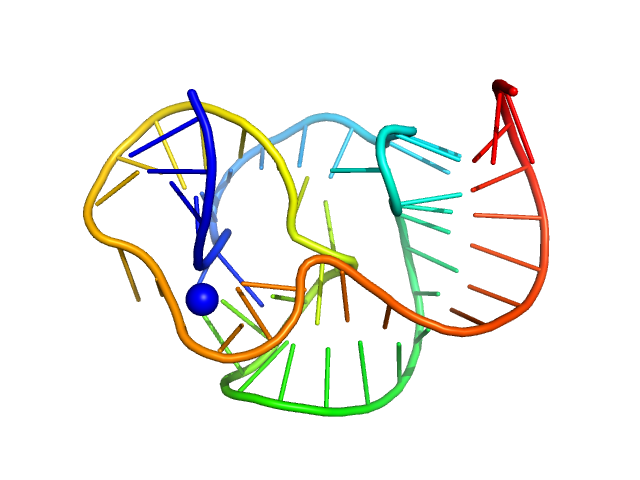
|
| Sample: |
ST9 WT (59 nt RNA) monomer, 19 kDa RNA
|
| Buffer: |
50 mM Tris pH 7.5, 100 mM NaCl, 1 mM DTT, 10 mM MgCl2, 0.0002% w/v NaN3, pH:
|
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2024 Jan 30
|
A conserved viral RNA fold enables nuclease resistance across kingdoms of life.
Nucleic Acids Res 53(16) (2025)
...Gong Z, Erickson A, Huang CH, Yang F, Cronin M, Kuo YW, Wimberly BT, Steckelberg AL
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
24 |
nm3 |
|