|
|
|
|
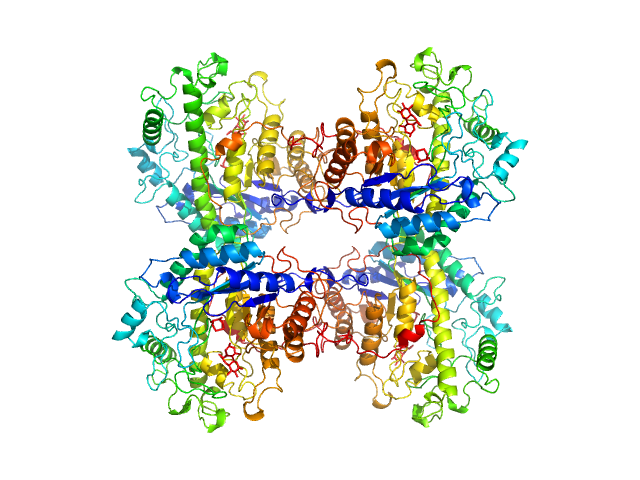
|
| Sample: |
Beta-amylase tetramer, 224 kDa Ipomoea batatas protein
|
| Buffer: |
phosphate buffered saline, 1% glycerol, pH: 7.2
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Apr 13
|
AF4-to-SAXS: expanded characterization of nanoparticles and proteins at the P12 BioSAXS beamline.
J Synchrotron Radiat (2025)
Da Vela S, Bartels K, Franke D, Soloviov D, Gräwert T, Molodenskiy D, Kolb B, Wilhelmy C, Drexel R, Meier F, Haas H, Langguth P, Graewert MA
|
| RgGuinier |
4.2 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
297 |
nm3 |
|