|
|
|
|
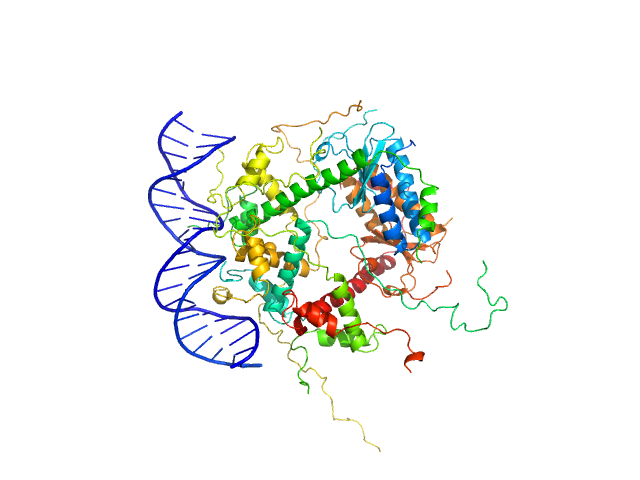
|
| Sample: |
Antitoxin ParD hexamer, 54 kDa Vibrio cholerae serotype … protein
Toxin , 25 kDa Vibrio cholerae serotype … protein
21-bp DNA operator fragment monomer, 13 kDa Vibrio cholerae O1 DNA
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 8
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Jul 18
|
Toxin:antitoxin ratio sensing autoregulation of the Vibrio cholerae parDE2 module.
Sci Adv 10(1):eadj2403 (2024)
Garcia-Rodriguez G, Girardin Y, Kumar Singh R, Volkov AN, Van Dyck J, Muruganandam G, Sobott F, Charlier D, Loris R
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
140 |
nm3 |
|