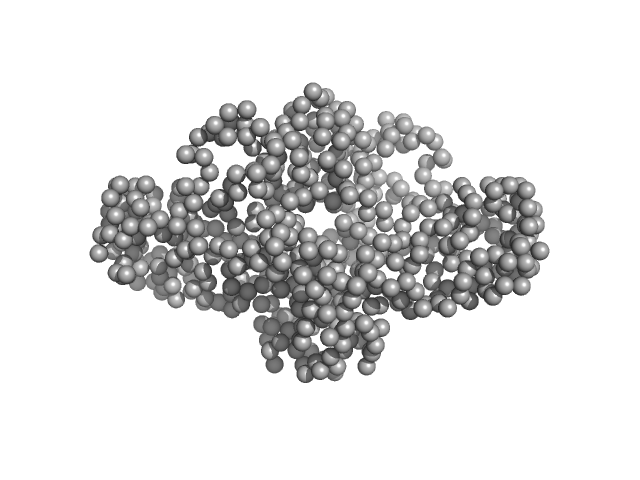
UniProt ID: A0A4Q5N6R9 (20-288) Uncharacterized protein
|
|
|
|
| Sample: |
Uncharacterized protein dimer, 66 kDa Legionella pneumophila protein
|
| Buffer: |
20 mM Tris, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Sep 18
|
Structure, Dynamics and Cellular Insight Into Novel Substrates of the Legionella pneumophila Type II Secretion System
Frontiers in Molecular Biosciences 7 (2020)
Portlock T, Tyson J, Dantu S, Rehman S, White R, McIntire I, Sewell L, Richardson K, Shaw R, Pandini A, Cianciotto N, Garnett J
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.4 |
nm |
| VolumePorod |
114 |
nm3 |
|
|
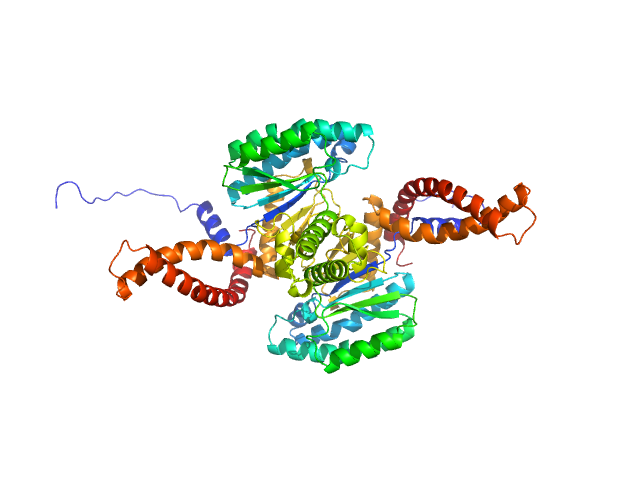
UniProt ID: Q86SQ9 (1-333) Dehydrodolichyl diphosphate synthase complex subunit DHDDS
|
|
|
|
| Sample: |
Dehydrodolichyl diphosphate synthase complex subunit DHDDS dimer, 78 kDa Homo sapiens protein
|
| Buffer: |
Tris-HCl, 150 mM NaCl, 20 mM 2-mercaptoethanol and 0.02% Triton-X100, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 2
|
Structural Characterization of Full-Length Human Dehydrodolichyl Diphosphate Synthase Using an Integrative Computational and Experimental Approach.
Biomolecules 9(11) (2019)
Lisnyansky Bar-El M, Lee SY, Ki AY, Kapelushnik N, Loewenstein A, Chung KY, Schneidman-Duhovny D, Giladi M, Newman H, Haitin Y
|
| RgGuinier |
3.2 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
122 |
nm3 |
|
|
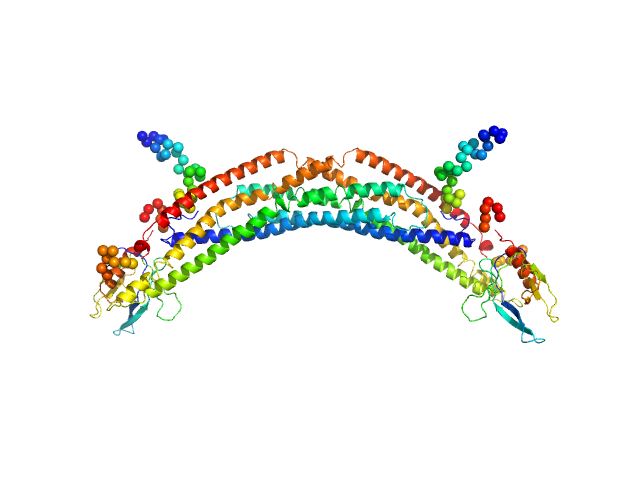
UniProt ID: Q8NEU8 (1-384) Adaptor protein, phosphotyrosine interaction, pleckstrin homology domain, and leucine zipper-containing protein 2
|
|
|
|
| Sample: |
Adaptor protein, phosphotyrosine interaction, pleckstrin homology domain, and leucine zipper-containing protein 2 dimer, 87 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, pH: 8.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 7 Apr 20
|
Membrane curvature protein exhibits interdomain flexibility and binds a small GTPase.
J Biol Chem 287(49):40996-1006 (2012)
King GJ, Stöckli J, Hu SH, Winnen B, Duprez WG, Meoli CC, Junutula JR, Jarrott RJ, James DE, Whitten AE, Martin JL
|
| RgGuinier |
5.1 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
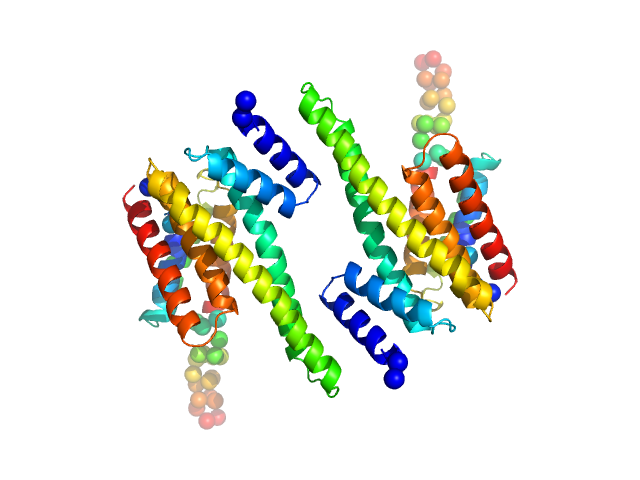
UniProt ID: P31946 (1-246) 14-3-3 protein beta/alpha
|
|
|
|
| Sample: |
14-3-3 protein beta/alpha dimer, 58 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, and 2 mM 2-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2011 Apr 7
|
The weak complex between RhoGAP protein ARHGAP22 and signal regulatory protein 14-3-3 has 1:2 stoichiometry and a single peptide binding mode.
PLoS One 7(8):e41731 (2012)
Hu SH, Whitten AE, King GJ, Jones A, Rowland AF, James DE, Martin JL
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
UniProt ID: None (53-551) Alpha domain of Ag43a
|
|
|

|
| Sample: |
Alpha domain of Ag43a monomer, 49 kDa Escherichia coli protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2009 Nov 19
|
The antigen 43 structure reveals a molecular Velcro-like mechanism of autotransporter-mediated bacterial clumping.
Proc Natl Acad Sci U S A 111(1):457-62 (2014)
Heras B, Totsika M, Peters KM, Paxman JJ, Gee CL, Jarrott RJ, Perugini MA, Whitten AE, Schembri MA
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
62 |
nm3 |
|
|
UniProt ID: Q7Z5H3 (1-405) Rho GTPase-activating protein 22
|
|
|
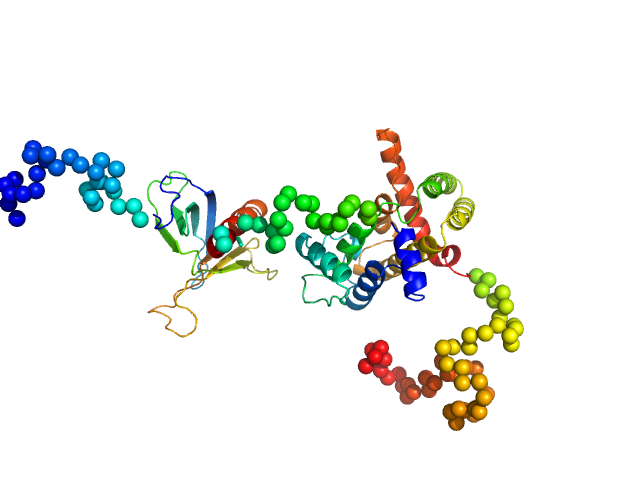
|
| Sample: |
Rho GTPase-activating protein 22 monomer, 47 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, and 2 mM 2-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2011 Apr 7
|
The weak complex between RhoGAP protein ARHGAP22 and signal regulatory protein 14-3-3 has 1:2 stoichiometry and a single peptide binding mode.
PLoS One 7(8):e41731 (2012)
Hu SH, Whitten AE, King GJ, Jones A, Rowland AF, James DE, Martin JL
|
| RgGuinier |
3.2 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
88 |
nm3 |
|
|
UniProt ID: P11881 (1-604) N-terminal domains of the inositol 1,4,5-trisphosphate receptor type 1
|
|
|
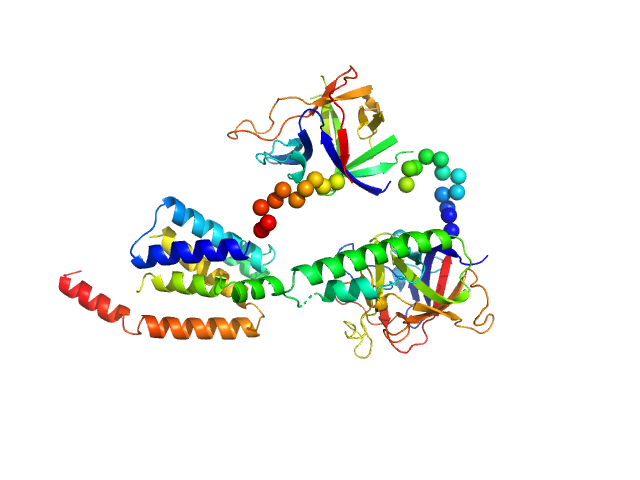
|
| Sample: |
N-terminal domains of the inositol 1,4,5-trisphosphate receptor type 1 monomer, 70 kDa Mus musculus protein
|
| Buffer: |
15 mM Tris, 300 mM NaCl, 1 mM TCEP, 5 mM EGTA, pH: 8 |
| Experiment: |
SAXS
data collected at Bruker Nanostar II, Australian Nuclear Science and Technology Organisation/Australian Centre for Neutron Scattering on 2006 Apr 12
|
Ligand-induced conformational changes via flexible linkers in the amino-terminal region of the inositol 1,4,5-trisphosphate receptor.
J Mol Biol 373(5):1269-80 (2007)
Chan J, Whitten AE, Jeffries CM, Bosanac I, Mal TK, Ito J, Porumb H, Michikawa T, Mikoshiba K, Trewhella J, Ikura M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
135 |
nm3 |
|
|
UniProt ID: P11881 (1-604) N-terminal domains of the inositol 1,4,5-trisphosphate receptor type 1
|
|
|
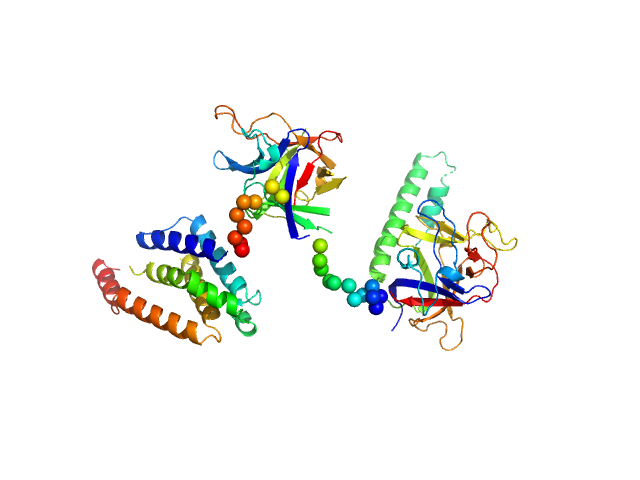
|
| Sample: |
N-terminal domains of the inositol 1,4,5-trisphosphate receptor type 1 monomer, 70 kDa Mus musculus protein
|
| Buffer: |
15 mM Tris, 300 mM NaCl, 1 mM TCEP, 10 mM CaCl2, pH: 8 |
| Experiment: |
SAXS
data collected at Bruker Nanostar II, Australian Nuclear Science and Technology Organisation/Australian Centre for Neutron Scattering on 2006 Apr 12
|
Ligand-induced conformational changes via flexible linkers in the amino-terminal region of the inositol 1,4,5-trisphosphate receptor.
J Mol Biol 373(5):1269-80 (2007)
Chan J, Whitten AE, Jeffries CM, Bosanac I, Mal TK, Ito J, Porumb H, Michikawa T, Mikoshiba K, Trewhella J, Ikura M
|
| RgGuinier |
3.4 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
160 |
nm3 |
|
|
UniProt ID: P11881 (1-604) N-terminal domains of the inositol 1,4,5-trisphosphate receptor type 1
|
|
|
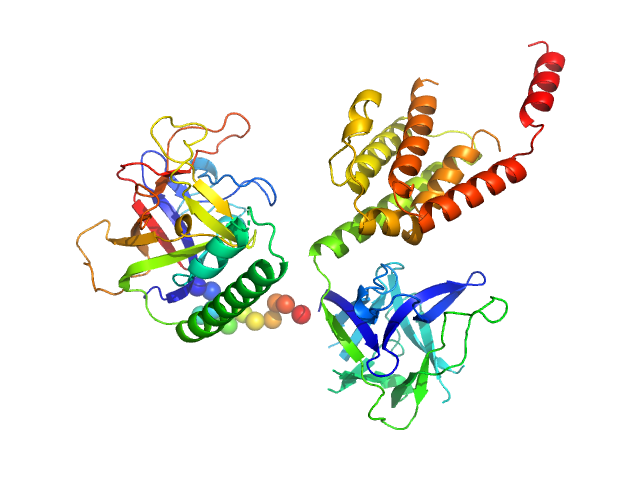
|
| Sample: |
N-terminal domains of the inositol 1,4,5-trisphosphate receptor type 1 monomer, 70 kDa Mus musculus protein
|
| Buffer: |
15 mM Tris, 300 mM NaCl, 1 mM TCEP, 5 mM EGTA, 0.25 mM IP3, pH: 8 |
| Experiment: |
SAXS
data collected at Bruker Nanostar II, Australian Nuclear Science and Technology Organisation/Australian Centre for Neutron Scattering on 2006 Apr 12
|
Ligand-induced conformational changes via flexible linkers in the amino-terminal region of the inositol 1,4,5-trisphosphate receptor.
J Mol Biol 373(5):1269-80 (2007)
Chan J, Whitten AE, Jeffries CM, Bosanac I, Mal TK, Ito J, Porumb H, Michikawa T, Mikoshiba K, Trewhella J, Ikura M
|
| RgGuinier |
3.1 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
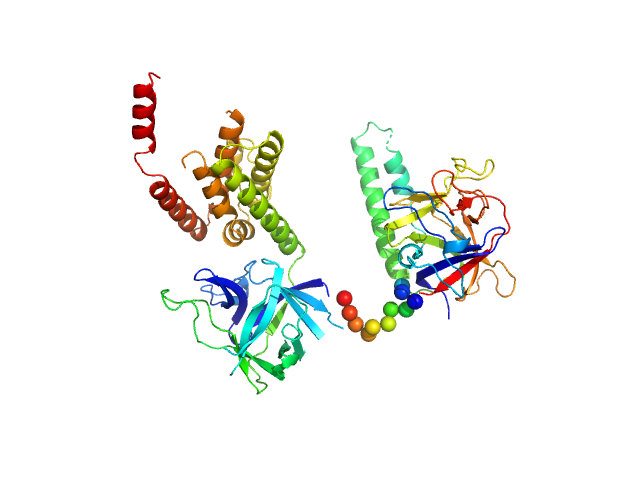
UniProt ID: P11881 (1-604) N-terminal domains of the inositol 1,4,5-trisphosphate receptor type 1
|
|
|
|
| Sample: |
N-terminal domains of the inositol 1,4,5-trisphosphate receptor type 1 monomer, 70 kDa Mus musculus protein
|
| Buffer: |
15 mM Tris, 300 mM NaCl, 1 mM TCEP, 10 mM CaCl2, 0.25 mM IP3, pH: 8 |
| Experiment: |
SAXS
data collected at Bruker Nanostar II, Australian Nuclear Science and Technology Organisation/Australian Centre for Neutron Scattering on 2006 Apr 12
|
Ligand-induced conformational changes via flexible linkers in the amino-terminal region of the inositol 1,4,5-trisphosphate receptor.
J Mol Biol 373(5):1269-80 (2007)
Chan J, Whitten AE, Jeffries CM, Bosanac I, Mal TK, Ito J, Porumb H, Michikawa T, Mikoshiba K, Trewhella J, Ikura M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
132 |
nm3 |
|
|