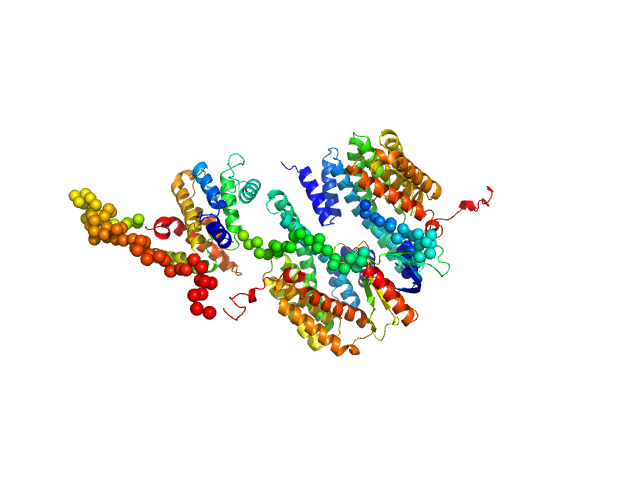
UniProt ID: P31946 (1-246) 14-3-3 protein beta/alpha
UniProt ID: Q7Z5H3 (1-405) Rho GTPase-activating protein 22
|
|
|
|
| Sample: |
14-3-3 protein beta/alpha dimer, 58 kDa Homo sapiens protein
Rho GTPase-activating protein 22 monomer, 47 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, and 2 mM 2-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2011 Apr 7
|
The weak complex between RhoGAP protein ARHGAP22 and signal regulatory protein 14-3-3 has 1:2 stoichiometry and a single peptide binding mode.
PLoS One 7(8):e41731 (2012)
Hu SH, Whitten AE, King GJ, Jones A, Rowland AF, James DE, Martin JL
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
195 |
nm3 |
|
|
UniProt ID: None (None-None) Braveheart RNA
UniProt ID: P62633 (1-177) Cellular nucleic acid-binding protein
|
|
|
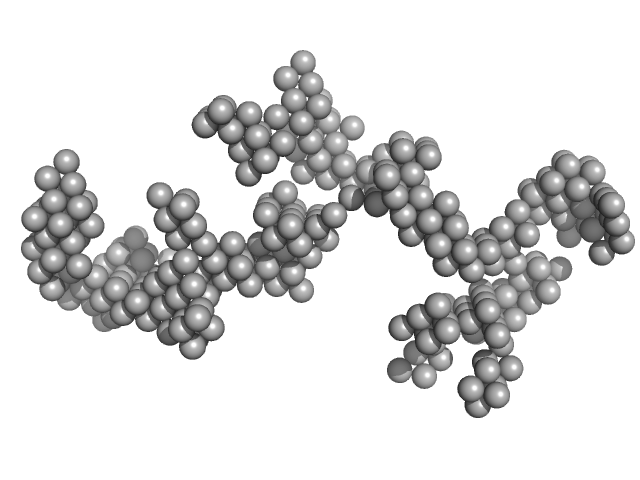
|
| Sample: |
Braveheart RNA monomer, 205 kDa Homo sapiens RNA
Cellular nucleic acid-binding protein monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES-KOH, 100 mM KCl, 6 mM MgCl2, pH: 7.6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jun 23
|
Zinc-finger protein CNBP alters the 3-D structure of lncRNA Braveheart in solution
Nature Communications 11(1) (2020)
Kim D, Thiel B, Mrozowich T, Hennelly S, Hofacker I, Patel T, Sanbonmatsu K
|
| RgGuinier |
9.8 |
nm |
| Dmax |
30.2 |
nm |
| VolumePorod |
1660 |
nm3 |
|
|
UniProt ID: P62633 (1-177) Cellular nucleic acid-binding protein
UniProt ID: None (None-None) Braveheart Fragment 1
|
|
|

|
| Sample: |
Cellular nucleic acid-binding protein monomer, 22 kDa Homo sapiens protein
Braveheart Fragment 1 monomer, 116 kDa Homo sapiens RNA
|
| Buffer: |
50 mM HEPES-KOH, 100 mM KCl, 6 mM MgCl2, pH: 7.6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jun 23
|
Zinc-finger protein CNBP alters the 3-D structure of lncRNA Braveheart in solution
Nature Communications 11(1) (2020)
Kim D, Thiel B, Mrozowich T, Hennelly S, Hofacker I, Patel T, Sanbonmatsu K
|
| RgGuinier |
8.2 |
nm |
| Dmax |
27.0 |
nm |
| VolumePorod |
455 |
nm3 |
|
|
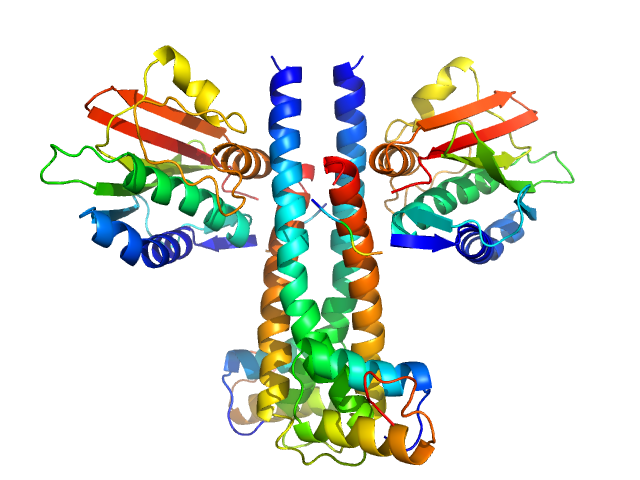
UniProt ID: P16497 (383-606) Sporulation kinase A
UniProt ID: Q7WY62 (7-52) Sporulation inhibitor sda
|
|
|
|
| Sample: |
Sporulation kinase A dimer, 51 kDa Bacillus subtilis protein
Sporulation inhibitor sda dimer, 11 kDa Bacillus subtilis protein
|
| Buffer: |
50mM Tris, 200mM NaCl, 150mM Imidazole, pH: 8.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar II, Australian Nuclear Science and Technology Organisation/Australian Centre for Neutron Scattering on 2006 Nov 16
|
The structure of the KinA-Sda complex suggests an allosteric mechanism of histidine kinase inhibition.
J Mol Biol 368(2):407-20 (2007)
Whitten AE, Jacques DA, Hammouda B, Hanley T, King GF, Guss JM, Trewhella J, Langley DB
|
| RgGuinier |
2.9 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
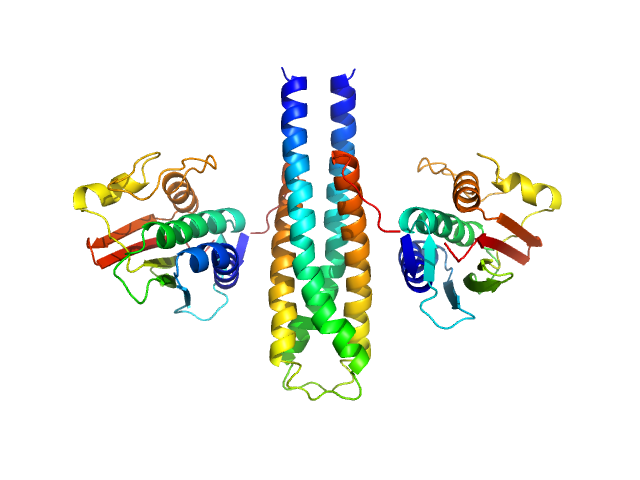
UniProt ID: P16497 (383-606) Sporulation kinase A
|
|
|
|
| Sample: |
Sporulation kinase A dimer, 51 kDa Bacillus subtilis protein
|
| Buffer: |
50mM Tris, 200mM NaCl, 150mM Imidazole, pH: 8.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar II, Australian Nuclear Science and Technology Organisation/Australian Centre for Neutron Scattering on 2006 Mar 18
|
The structure of the KinA-Sda complex suggests an allosteric mechanism of histidine kinase inhibition.
J Mol Biol 368(2):407-20 (2007)
Whitten AE, Jacques DA, Hammouda B, Hanley T, King GF, Guss JM, Trewhella J, Langley DB
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
UniProt ID: A8K7I4 (302-476) Calcium-activated chloride channel regulator 1
|
|
|

|
| Sample: |
Calcium-activated chloride channel regulator 1 monomer, 20 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Jun 6
|
Structural and Biophysical Analysis of the CLCA1 VWA Domain Suggests Mode of TMEM16A Engagement.
Cell Rep 30(4):1141-1151.e3 (2020)
Berry KN, Brett TJ
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.6 |
nm |
|
|
UniProt ID: A8K7I4 (22-477) Calcium-activated chloride channel regulator 1
|
|
|

|
| Sample: |
Calcium-activated chloride channel regulator 1 monomer, 52 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Jun 6
|
Structural and Biophysical Analysis of the CLCA1 VWA Domain Suggests Mode of TMEM16A Engagement.
Cell Rep 30(4):1141-1151.e3 (2020)
Berry KN, Brett TJ
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.4 |
nm |
|
|
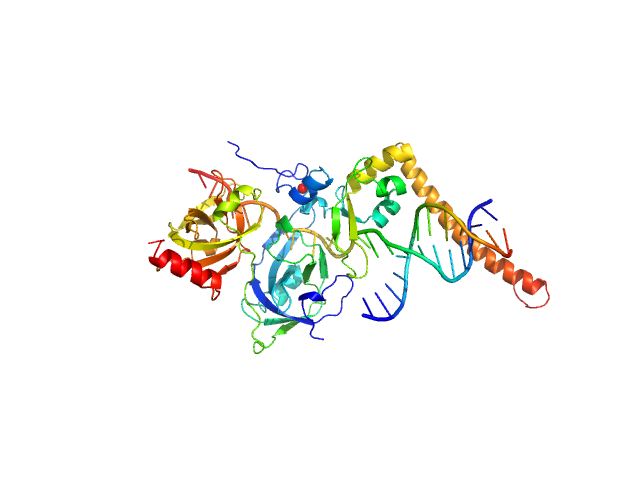
UniProt ID: P23025 (98-239) DNA repair protein complementing XP-A cells
UniProt ID: P27694 (183-420) Replication protein A 70 kDa DNA-binding subunit
UniProt ID: None (None-None) 3-prime Nucleotide Excision Repair Junction Model Substrate
|
|
|
|
| Sample: |
DNA repair protein complementing XP-A cells monomer, 17 kDa Homo sapiens protein
Replication protein A 70 kDa DNA-binding subunit monomer, 27 kDa Homo sapiens protein
3-prime Nucleotide Excision Repair Junction Model Substrate monomer, 11 kDa DNA
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2% glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 17 Nov 2
|
A key interaction with RPA orients XPA in NER complexes.
Nucleic Acids Res (2020)
Topolska-Woś AM, Sugitani N, Cordoba JJ, Le Meur KV, Le Meur RA, Kim HS, Yeo JE, Rosenberg D, Hammel M, Schärer OD, Chazin WJ
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
103 |
nm3 |
|
|
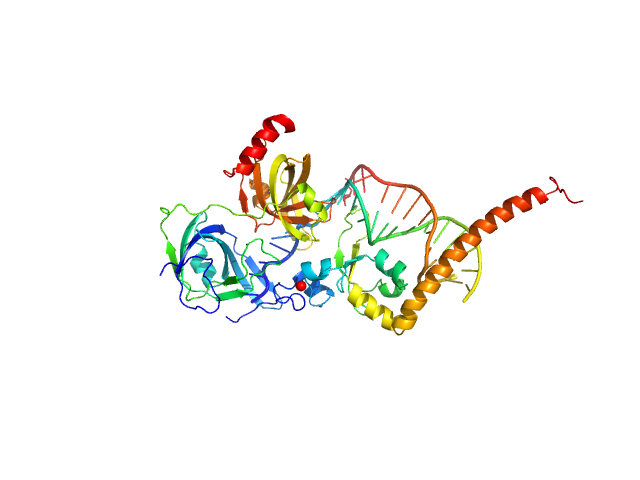
UniProt ID: P23025 (98-239) DNA repair protein complementing XP-A cells
UniProt ID: P27694 (183-420) Replication protein A 70 kDa DNA-binding subunit
UniProt ID: None (None-None) 5-prime Nucleotide Excision Repair Junction Model Substrate
|
|
|
|
| Sample: |
DNA repair protein complementing XP-A cells monomer, 17 kDa Homo sapiens protein
Replication protein A 70 kDa DNA-binding subunit monomer, 27 kDa Homo sapiens protein
5-prime Nucleotide Excision Repair Junction Model Substrate monomer, 11 kDa DNA
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2% glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Jun 4
|
A key interaction with RPA orients XPA in NER complexes.
Nucleic Acids Res (2020)
Topolska-Woś AM, Sugitani N, Cordoba JJ, Le Meur KV, Le Meur RA, Kim HS, Yeo JE, Rosenberg D, Hammel M, Schärer OD, Chazin WJ
|
| RgGuinier |
2.9 |
nm |
| Dmax |
97.0 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
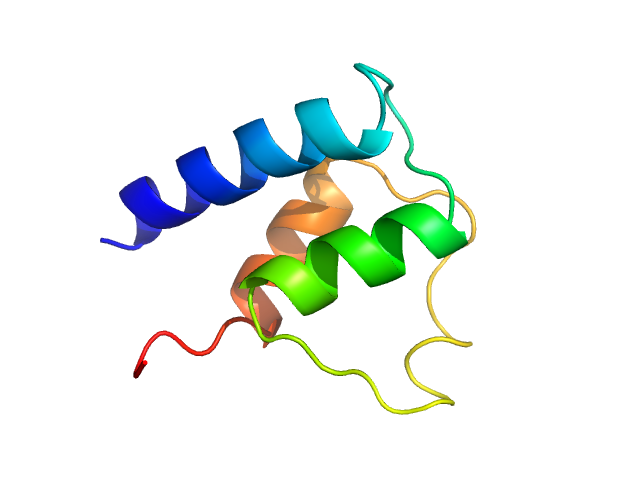
UniProt ID: Q8IJM4 (1-74) Myosin essential light chain
|
|
|
|
| Sample: |
Myosin essential light chain monomer, 9 kDa Plasmodium falciparum protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Oct 25
|
Structural role of essential light chains in the apicomplexan glideosome.
Commun Biol 3(1):568 (2020)
Pazicky S, Dhamotharan K, Kaszuba K, Mertens HDT, Gilberger T, Svergun D, Kosinski J, Weininger U, Löw C
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.3 |
nm |
| VolumePorod |
12 |
nm3 |
|
|