UniProt ID: Q9EQZ6-3 (1-993) Isoform 3 of Rap guanine nucleotide exchange factor 4
|
|
|
|
| Sample: |
Isoform 3 of Rap guanine nucleotide exchange factor 4 monomer, 114 kDa Mus musculus protein
|
| Buffer: |
150 mM NaCl, 1 mM EDTA, 1 mM DTT, and 10 mM Tris-HCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSess, University of Utah on 2008 Aug 7
|
Mechanism of Epac activation: structural and functional analyses of Epac2 hinge mutants with constitutive and reduced activities.
J Biol Chem 284(35):23644-51 (2009)
Tsalkova T, Blumenthal DK, Mei FC, White MA, Cheng X
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
123 |
nm3 |
|
|
UniProt ID: Q9EQZ6-3 (1-993) Isoform 3 of Rap guanine nucleotide exchange factor 4
|
|
|
|
| Sample: |
Isoform 3 of Rap guanine nucleotide exchange factor 4 monomer, 114 kDa Mus musculus protein
|
| Buffer: |
150 mM NaCl, 1 mM EDTA, 1 mM DTT, and 10 mM Tris-HCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSess, University of Utah on 2008 Aug 8
|
Mechanism of Epac activation: structural and functional analyses of Epac2 hinge mutants with constitutive and reduced activities.
J Biol Chem 284(35):23644-51 (2009)
Tsalkova T, Blumenthal DK, Mei FC, White MA, Cheng X
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
151 |
nm3 |
|
|
UniProt ID: Q9EQZ6-3 (1-993) Isoform 3 of Rap guanine nucleotide exchange factor 4
|
|
|
|
| Sample: |
Isoform 3 of Rap guanine nucleotide exchange factor 4 monomer, 114 kDa Mus musculus protein
|
| Buffer: |
150 mM NaCl, 1 mM EDTA, 1 mM DTT, and 10 mM Tris-HCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSess, University of Utah on 2008 Aug 11
|
Mechanism of Epac activation: structural and functional analyses of Epac2 hinge mutants with constitutive and reduced activities.
J Biol Chem 284(35):23644-51 (2009)
Tsalkova T, Blumenthal DK, Mei FC, White MA, Cheng X
|
| RgGuinier |
3.6 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
145 |
nm3 |
|
|
UniProt ID: Q8I2Q0 (22-217) Plasmodium falciparum Lipocalin
|
|
|
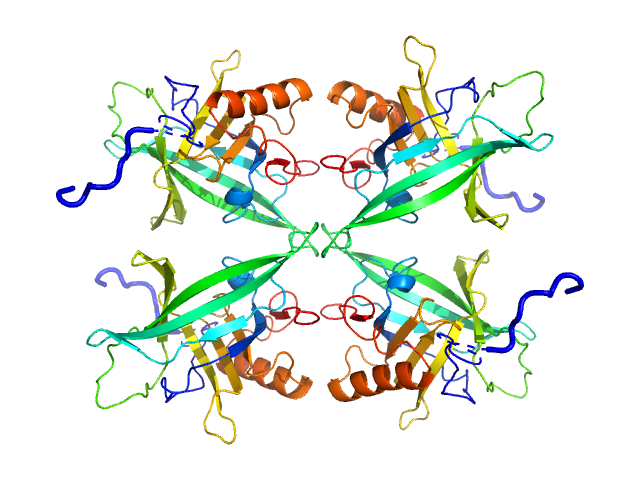
|
| Sample: |
Plasmodium falciparum Lipocalin tetramer, 89 kDa Plasmodium falciparum protein
|
| Buffer: |
20 mM Tris pH7.5, 150 mM NaCl, 5% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Apr 8
|
Structure-Based Identification and Functional Characterization of a Lipocalin in the Malaria Parasite Plasmodium falciparum
Cell Reports 31(12):107817 (2020)
Burda P, Crosskey T, Lauk K, Zurborg A, Söhnchen C, Liffner B, Wilcke L, Pietsch E, Strauss J, Jeffries C, Svergun D, Wilson D, Wilmanns M, Gilberger T
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
126 |
nm3 |
|
|
UniProt ID: O77105 (1-699) Soluble guanylyl cyclase alpha-1 subunit
UniProt ID: O77106 (1-600) Soluble guanylyl cyclase beta-1 subunit
|
|
|
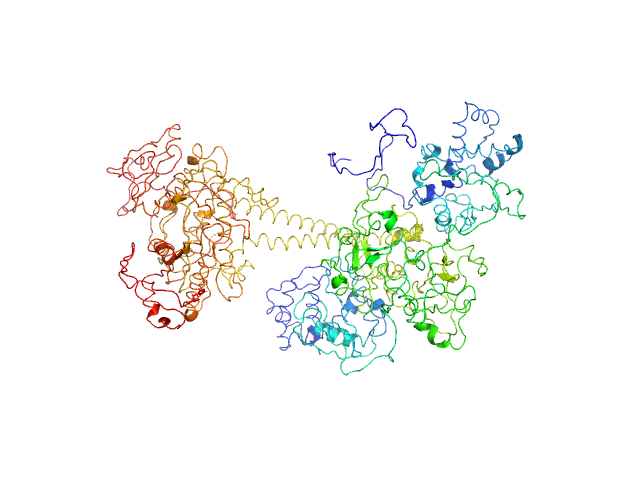
|
| Sample: |
Soluble guanylyl cyclase alpha-1 subunit monomer, 78 kDa Manduca sexta protein
Soluble guanylyl cyclase beta-1 subunit monomer, 68 kDa Manduca sexta protein
|
| Buffer: |
50 mM KH2PO4, 150 mM NaCl, 2% glycerol,, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Dec 3
|
Allosteric activation of the nitric oxide receptor soluble guanylate cyclase mapped by cryo-electron microscopy.
Elife 8 (2019)
Horst BG, Yokom AL, Rosenberg DJ, Morris KL, Hammel M, Hurley JH, Marletta MA
|
| RgGuinier |
4.3 |
nm |
| Dmax |
13.3 |
nm |
| VolumePorod |
230 |
nm3 |
|
|
UniProt ID: O77105 (1-699) Soluble guanylyl cyclase alpha-1 subunit
UniProt ID: O77106 (1-600) Soluble guanylyl cyclase beta-1 subunit
|
|
|
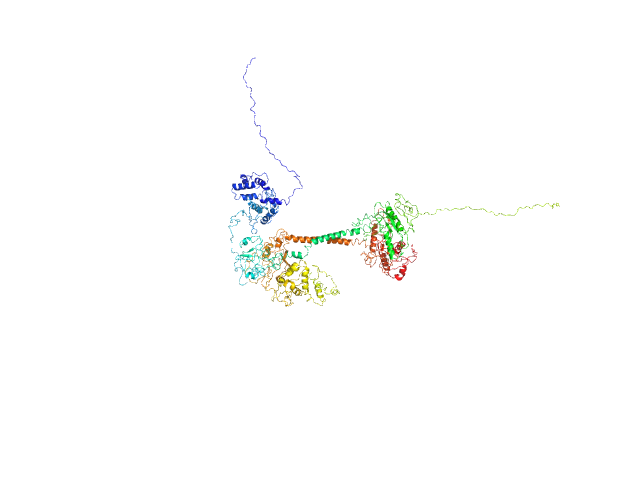
|
| Sample: |
Soluble guanylyl cyclase alpha-1 subunit monomer, 78 kDa Manduca sexta protein
Soluble guanylyl cyclase beta-1 subunit monomer, 68 kDa Manduca sexta protein
|
| Buffer: |
50 mM KH2PO4,150 mM NaCl, 2% glycerol, 500 µM NO,, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Dec 3
|
Allosteric activation of the nitric oxide receptor soluble guanylate cyclase mapped by cryo-electron microscopy.
Elife 8 (2019)
Horst BG, Yokom AL, Rosenberg DJ, Morris KL, Hammel M, Hurley JH, Marletta MA
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
218 |
nm3 |
|
|
UniProt ID: W0C4C9 (1-598) 2,4-dichlorophenol 6-monooxygenase
UniProt ID: None (None-None) Flavin adenine dinucleotide
|
|
|
|
| Sample: |
2,4-dichlorophenol 6-monooxygenase hexamer, 399 kDa Streptomyces sp. SCSIO … protein
Flavin adenine dinucleotide hexamer, 5 kDa
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 5 mM DTT, 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with MetalJet, Département de Biochimie, Université de Montréal on 2019 Oct 22
|
Structural analyses of the group A flavin-dependent monooxygenase PieE reveal a sliding FAD cofactor conformation bridging OUT and IN conformations.
J Biol Chem (2020)
Manenda MS, Picard MÈ, Zhang L, Cyr N, Zhu X, Barma J, Pascal JM, Couture M, Zhang C, Shi R
|
| RgGuinier |
4.8 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
624 |
nm3 |
|
|
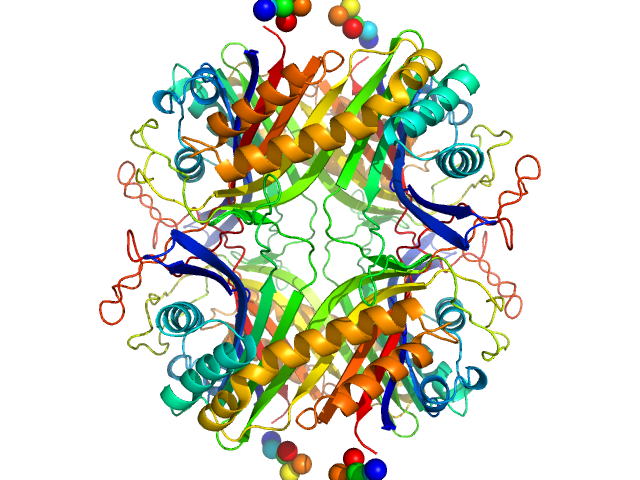
UniProt ID: Q00511 (1-302) Urate Oxidase (Uricase) from Aspergillus flavus
|
|
|
|
| Sample: |
Urate Oxidase (Uricase) from Aspergillus flavus tetramer, 137 kDa Aspergillus flavus protein
|
| Buffer: |
20 mM Tris. 150 mM NaCl, 1 mM EDTA, 5 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2019 Jul 1
|
Urate Oxidase (Uricase) from Aspergillus flavus
Tsutomu Matsui
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
225 |
nm3 |
|
|
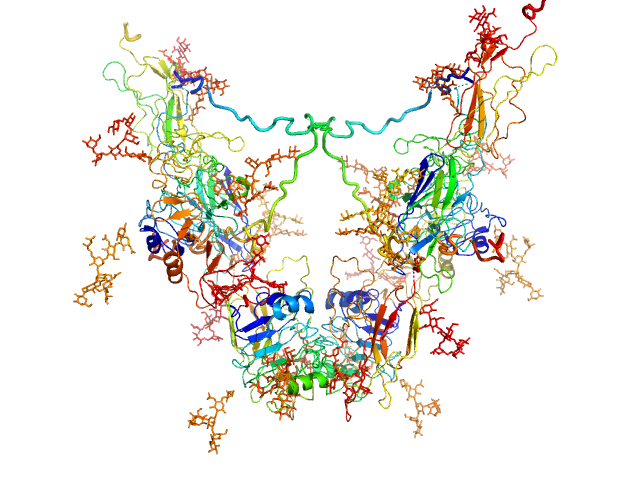
UniProt ID: P08069 (31-935) Insulin-like growth factor 1 receptor
|
|
|
|
| Sample: |
Insulin-like growth factor 1 receptor dimer, 202 kDa Homo sapiens protein
|
| Buffer: |
30 mM Tris, 140 mM NaCl, 0.02% w/v sodium azide,, pH: 7.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar, Australian Nuclear Science and Technology Organisation on 2008 Apr 22
|
Solution structure of ectodomains of the insulin receptor family: the ectodomain of the type 1 insulin-like growth factor receptor displays asymmetry of ligand binding accompanied by limited conformational change.
J Mol Biol 394(5):878-92 (2009)
Whitten AE, Smith BJ, Menting JG, Margetts MB, McKern NM, Lovrecz GO, Adams TE, Richards K, Bentley JD, Trewhella J, Ward CW, Lawrence MC
|
| RgGuinier |
5.1 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
357 |
nm3 |
|
|
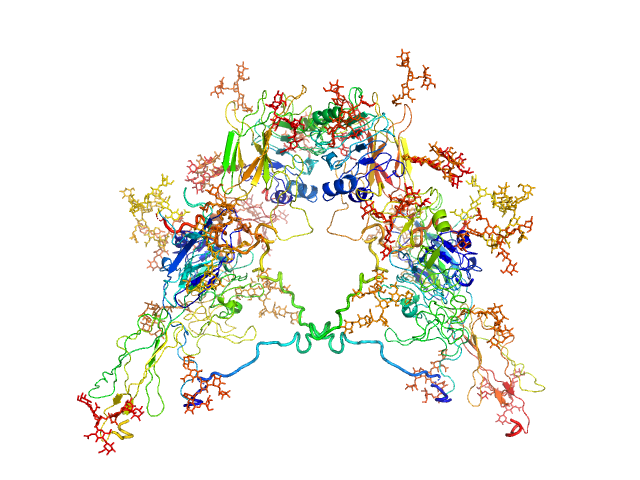
UniProt ID: P06213-1 (28-948) Insulin receptor
|
|
|
|
| Sample: |
Insulin receptor dimer, 203 kDa Homo sapiens protein
|
| Buffer: |
30 mM Tris, 140 mM NaCl, 0.02% w/v sodium azide,, pH: 7.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar, Australian Nuclear Science and Technology Organisation on 2008 Dec 7
|
Solution structure of ectodomains of the insulin receptor family: the ectodomain of the type 1 insulin-like growth factor receptor displays asymmetry of ligand binding accompanied by limited conformational change.
J Mol Biol 394(5):878-92 (2009)
Whitten AE, Smith BJ, Menting JG, Margetts MB, McKern NM, Lovrecz GO, Adams TE, Richards K, Bentley JD, Trewhella J, Ward CW, Lawrence MC
|
| RgGuinier |
5.5 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
385 |
nm3 |
|
|