UniProt ID: F2L613 (1-401) Trehalose transferase (Trehalose phosphorylase/synthase)
|
|
|

|
| Sample: |
Trehalose transferase (Trehalose phosphorylase/synthase) monomer, 47 kDa Thermoproteus uzoniensis protein
|
| Buffer: |
50 mM HEPES, 100 NaCl, 4 mM DTT, 20 mM MgCl2, 1 mM UDP-glucose, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 27
|
Anomeric Selectivity of Trehalose Transferase with Rare L-Sugars
ACS Catalysis (2020)
Mestrom L, Marsden S, van der Eijk H, Laustsen J, Jeffries C, Svergun D, Hagedoorn P, Bento I, Hanefeld U
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
73 |
nm3 |
|
|
UniProt ID: Q5AMN0 (1-259) Proliferating cell nuclear antigen (C-terminal His-tagged)
|
|
|

|
| Sample: |
Proliferating cell nuclear antigen (C-terminal His-tagged) trimer, 90 kDa Candida albicans protein
|
| Buffer: |
20 mM Tris-HCl (pH 7.5), 100 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Sep 3
|
Structural analyses of PCNA from the fungal pathogen Candida albicans identify three regions with species-specific conformations.
FEBS Lett (2021)
Sundaram R, Manohar K, Patel SK, Acharya N, Vasudevan D
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
128 |
nm3 |
|
|
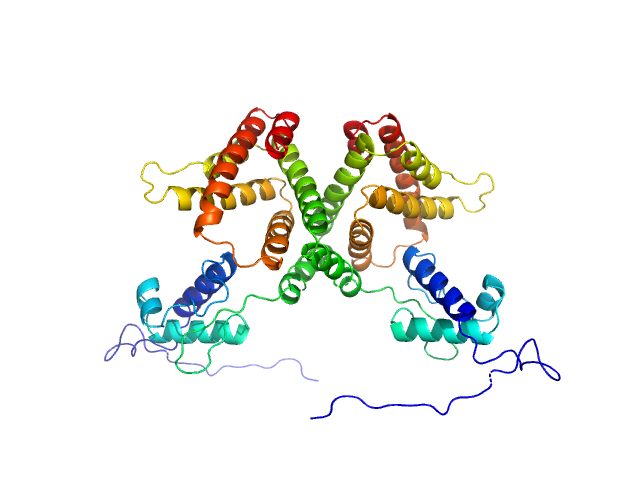
UniProt ID: P0A8W0 (1-263) HTH-type transcriptional repressor NanR
|
|
|
|
| Sample: |
HTH-type transcriptional repressor NanR dimer, 59 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.1 % (w/v) sodium azide, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Apr 20
|
Mechanism of NanR gene repression and allosteric induction of bacterial sialic acid metabolism
(2020)
Horne C, Venugopal H, Panjikar S, Henrickson A, Brookes E, North R, Murphy J, Friemann R, Griffin M, Ramm G, Demeler B, Dobson R
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
110 |
nm3 |
|
|
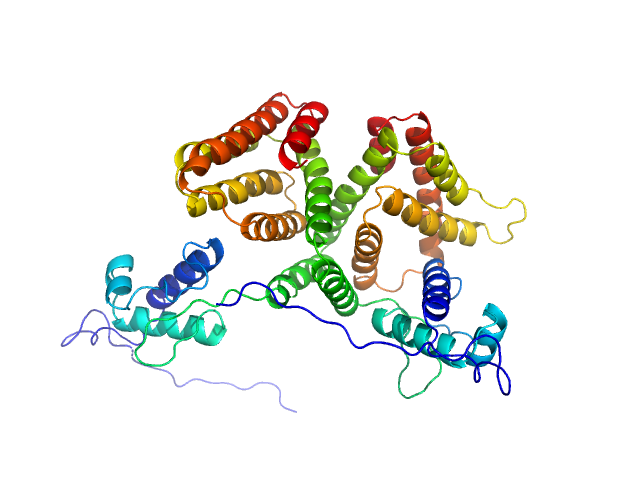
UniProt ID: P0A8W0 (1-263) HTH-type transcriptional repressor NanR
|
|
|
|
| Sample: |
HTH-type transcriptional repressor NanR dimer, 59 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 20 mM Neu5Ac and 0.1 % (w/v) sodium azide, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Apr 20
|
Mechanism of NanR gene repression and allosteric induction of bacterial sialic acid metabolism
(2020)
Horne C, Venugopal H, Panjikar S, Henrickson A, Brookes E, North R, Murphy J, Friemann R, Griffin M, Ramm G, Demeler B, Dobson R
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
105 |
nm3 |
|
|
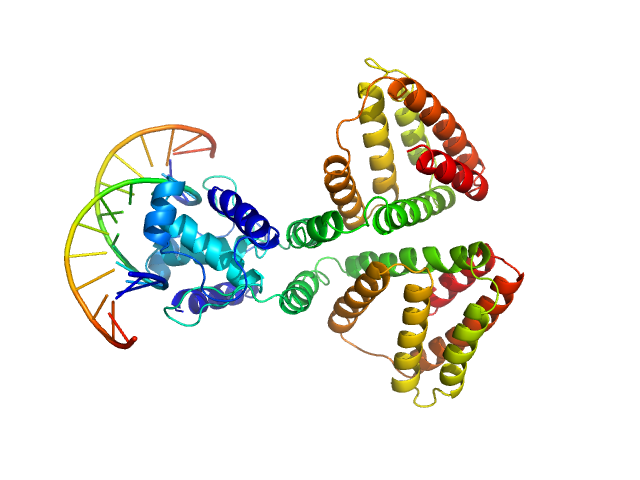
UniProt ID: P0A8W0 (1-263) HTH-type transcriptional repressor NanR
UniProt ID: None (None-None) (GGTATA)2 repeat DNA
|
|
|
|
| Sample: |
HTH-type transcriptional repressor NanR dimer, 59 kDa Escherichia coli protein
(GGTATA)2 repeat DNA monomer, 11 kDa DNA
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.1 % (w/v) sodium azide, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Apr 20
|
Mechanism of NanR gene repression and allosteric induction of bacterial sialic acid metabolism
(2020)
Horne C, Venugopal H, Panjikar S, Henrickson A, Brookes E, North R, Murphy J, Friemann R, Griffin M, Ramm G, Demeler B, Dobson R
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
108 |
nm3 |
|
|
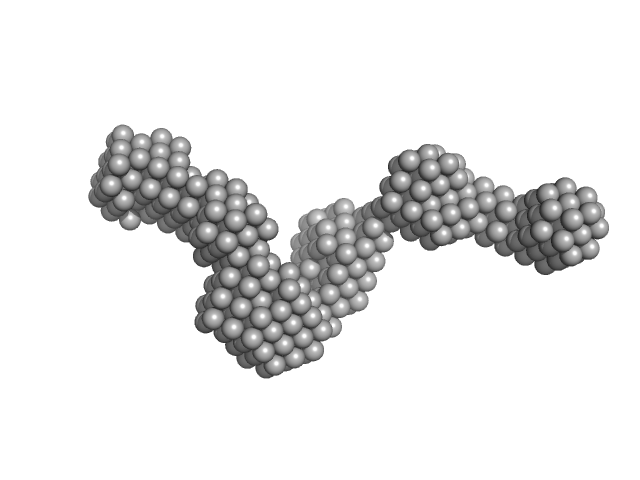
UniProt ID: A5FB63 (464-1137) Chitinase ChiA
|
|
|
|
| Sample: |
Chitinase ChiA monomer, 70 kDa Flavobacterium johnsoniae protein
|
| Buffer: |
50 mM Tris, 250 mM NaCl, 0.25 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2019 May 7
|
Structural insights of the enzymes from the chitin utilization locus of Flavobacterium johnsoniae.
Sci Rep 10(1):13775 (2020)
Mazurkewich S, Helland R, Mackenzie A, Eijsink VGH, Pope PB, Brändén G, Larsbrink J
|
| RgGuinier |
4.7 |
nm |
| Dmax |
16.3 |
nm |
| VolumePorod |
99 |
nm3 |
|
|
UniProt ID: P04925 (23-230) Major prion protein
|
|
|
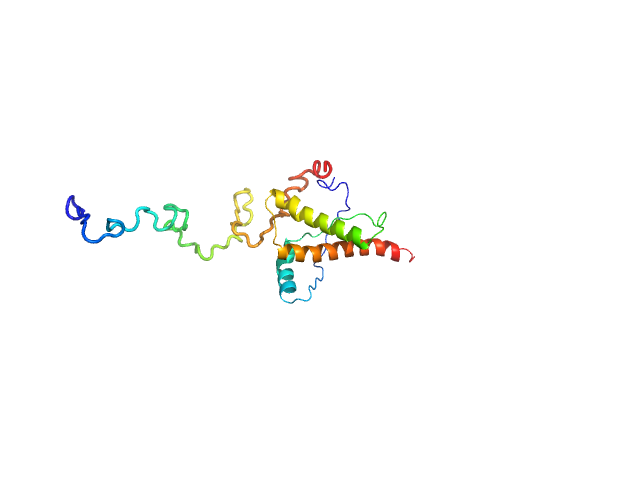
|
| Sample: |
Major prion protein monomer, 23 kDa Mus musculus protein
|
| Buffer: |
10mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 15
|
Characterization of murine prion protein
Stefano Da Vela
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
UniProt ID: G0SB82 (3-1222) Condensin complex subunit 1
|
|
|
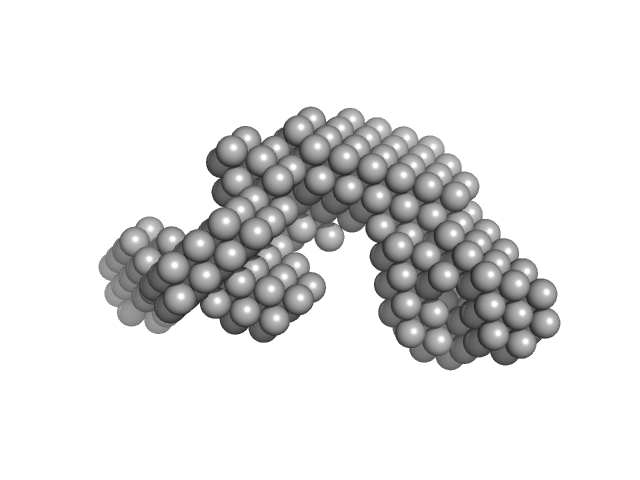
|
| Sample: |
Condensin complex subunit 1 monomer, 137 kDa Chaetomium thermophilum protein
|
| Buffer: |
25 mM Tris, 300 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Nov 20
|
Molecular flexibility of the condensin subunit Ycs4 is modulated by kleisin binding
Karen Manalastas-Cantos
|
| RgGuinier |
5.2 |
nm |
| Dmax |
17.7 |
nm |
| VolumePorod |
296 |
nm3 |
|
|
UniProt ID: G0SB82 (3-1222) Condensin complex subunit 1
UniProt ID: G0SBJ6 (336-418) Condensin complex subunit 2, 336-418
|
|
|
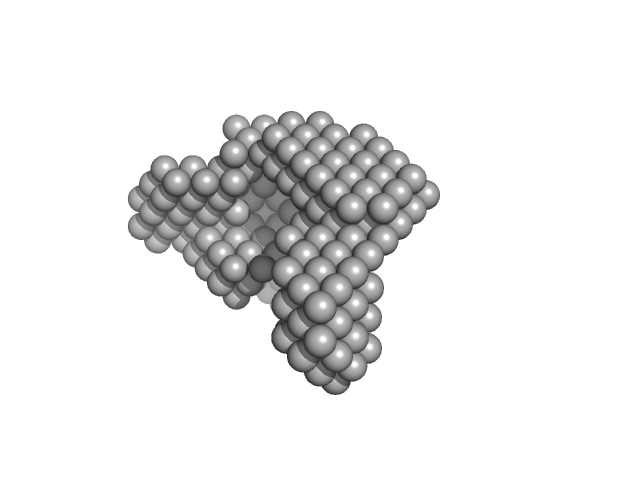
|
| Sample: |
Condensin complex subunit 1 monomer, 137 kDa Chaetomium thermophilum protein
Condensin complex subunit 2, 336-418 monomer, 9 kDa Chaetomium thermophilum protein
|
| Buffer: |
25 mM Tris, 300 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Nov 20
|
Molecular flexibility of the condensin subunit Ycs4 is modulated by kleisin binding
Karen Manalastas-Cantos
|
| RgGuinier |
5.0 |
nm |
| Dmax |
16.9 |
nm |
| VolumePorod |
279 |
nm3 |
|
|
UniProt ID: G0SB82 (3-1222) Condensin complex subunit 1
UniProt ID: G0SBJ6 (225-418) Condensin complex subunit 2, 225-418
|
|
|
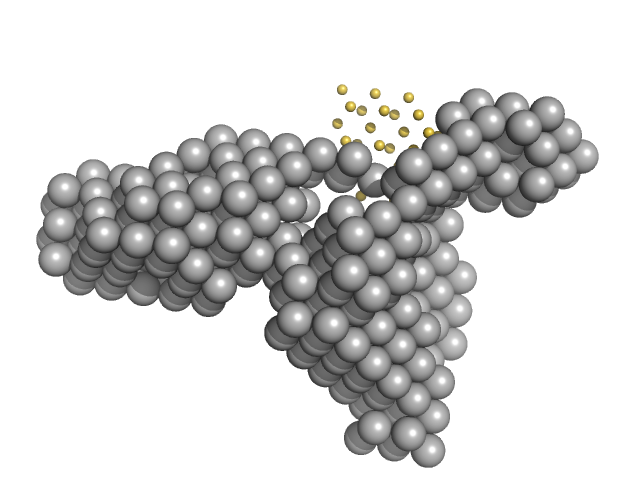
|
| Sample: |
Condensin complex subunit 1 monomer, 137 kDa Chaetomium thermophilum protein
Condensin complex subunit 2, 225-418 monomer, 21 kDa Chaetomium thermophilum protein
|
| Buffer: |
25 mM Tris, 300 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Nov 20
|
Molecular flexibility of the condensin subunit Ycs4 is modulated by kleisin binding
Karen Manalastas-Cantos
|
| RgGuinier |
4.9 |
nm |
| Dmax |
15.9 |
nm |
| VolumePorod |
309 |
nm3 |
|
|