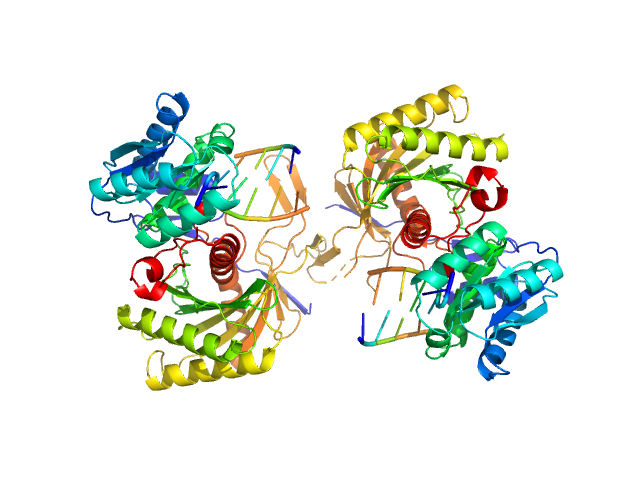
UniProt ID: O28951 (None-None) Piwi protein AF_1318
UniProt ID: None (None-None) 5'-phosphorilated 14-mer DNA oligoduplex
|
|
|
|
| Sample: |
Piwi protein AF_1318 dimer, 102 kDa Archaeoglobus fulgidus protein
5'-phosphorilated 14-mer DNA oligoduplex dimer, 9 kDa DNA
|
| Buffer: |
20 mM TrisHCl, pH 7.5, 5 mM MgCl2, 150 mM NaCl and 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 6
|
Prokaryotic Argonaute from Archaeoglobus fulgidus interacts with DNA as a homodimer.
Sci Rep 11(1):4518 (2021)
Golovinas E, Rutkauskas D, Manakova E, Jankunec M, Silanskas A, Sasnauskas G, Zaremba M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
158 |
nm3 |
|
|
UniProt ID: E9QNR8 (514-595) BRME1
UniProt ID: A0A3T0ZJB3 (18-122) Meiotic localizer of BRCA2
|
|
|

|
| Sample: |
BRME1 dimer, 23 kDa Mus musculus protein
Meiotic localizer of BRCA2 dimer, 26 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Oct 7
|
The BRCA2-MEILB2-BRME1 complex governs meiotic recombination and impairs the mitotic BRCA2-RAD51 function in cancer cells
Nature Communications 11(1) (2020)
Zhang J, Gurusaran M, Fujiwara Y, Zhang K, Echbarthi M, Vorontsov E, Guo R, Pendlebury D, Alam I, Livera G, Emmanuelle M, Wang P, Nandakumar J, Davies O, Shibuya H
|
| RgGuinier |
5.0 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
104 |
nm3 |
|
|
UniProt ID: A0A3T0ZJB3 (18-122) Meiotic localizer of BRCA2
|
|
|

|
| Sample: |
Meiotic localizer of BRCA2 dimer, 26 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Oct 7
|
The BRCA2-MEILB2-BRME1 complex governs meiotic recombination and impairs the mitotic BRCA2-RAD51 function in cancer cells
Nature Communications 11(1) (2020)
Zhang J, Gurusaran M, Fujiwara Y, Zhang K, Echbarthi M, Vorontsov E, Guo R, Pendlebury D, Alam I, Livera G, Emmanuelle M, Wang P, Nandakumar J, Davies O, Shibuya H
|
| RgGuinier |
4.6 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
88 |
nm3 |
|
|
UniProt ID: A0A3T0ZJB3 (18-122) Meiotic localizer of BRCA2
|
|
|

|
| Sample: |
Meiotic localizer of BRCA2 octamer, 102 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Oct 7
|
The BRCA2-MEILB2-BRME1 complex governs meiotic recombination and impairs the mitotic BRCA2-RAD51 function in cancer cells
Nature Communications 11(1) (2020)
Zhang J, Gurusaran M, Fujiwara Y, Zhang K, Echbarthi M, Vorontsov E, Guo R, Pendlebury D, Alam I, Livera G, Emmanuelle M, Wang P, Nandakumar J, Davies O, Shibuya H
|
| RgGuinier |
5.7 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
368 |
nm3 |
|
|
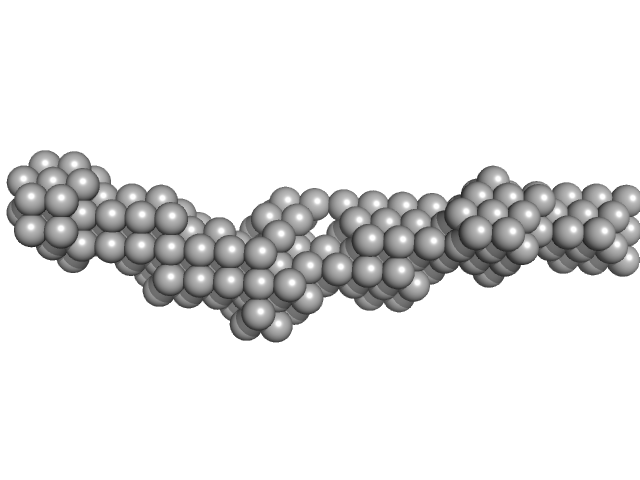
UniProt ID: A0A3T0ZJB3 (51-122) Meiotic localizer of BRCA2
|
|
|
|
| Sample: |
Meiotic localizer of BRCA2 monomer, 9 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Oct 7
|
The BRCA2-MEILB2-BRME1 complex governs meiotic recombination and impairs the mitotic BRCA2-RAD51 function in cancer cells
Nature Communications 11(1) (2020)
Zhang J, Gurusaran M, Fujiwara Y, Zhang K, Echbarthi M, Vorontsov E, Guo R, Pendlebury D, Alam I, Livera G, Emmanuelle M, Wang P, Nandakumar J, Davies O, Shibuya H
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
UniProt ID: Q8IC05 (1-745) Plasmodium falciparum Heat shock protein 90
|
|
|

|
| Sample: |
Plasmodium falciparum Heat shock protein 90 dimer, 177 kDa Plasmodium falciparum protein
|
| Buffer: |
25 mM Tris-HCl, 100 mM KCl, 1 mM β-mercaptoethanol, 1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS1 Beamline, Brazilian Synchrotron Light Laboratory on 2018 May 11
|
Solution structure of Plasmodium falciparum Hsp90 indicates a high flexible dimer
Archives of Biochemistry and Biophysics :108468 (2020)
Silva N, Torricillas M, Minari K, Barbosa L, Seraphim T, Borges J
|
| RgGuinier |
5.7 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
350 |
nm3 |
|
|
UniProt ID: Q8IC05 (312-566) Plasmodium falciparum Heat shock protein 90 middle domain
|
|
|

|
| Sample: |
Plasmodium falciparum Heat shock protein 90 middle domain monomer, 33 kDa Plasmodium falciparum protein
|
| Buffer: |
25 mM Tris-HCl, 100 mM KCl, 1 mM β-mercaptoethanol, 1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS1 Beamline, Brazilian Synchrotron Light Laboratory on 2018 May 11
|
Solution structure of Plasmodium falciparum Hsp90 indicates a high flexible dimer
Archives of Biochemistry and Biophysics :108468 (2020)
Silva N, Torricillas M, Minari K, Barbosa L, Seraphim T, Borges J
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
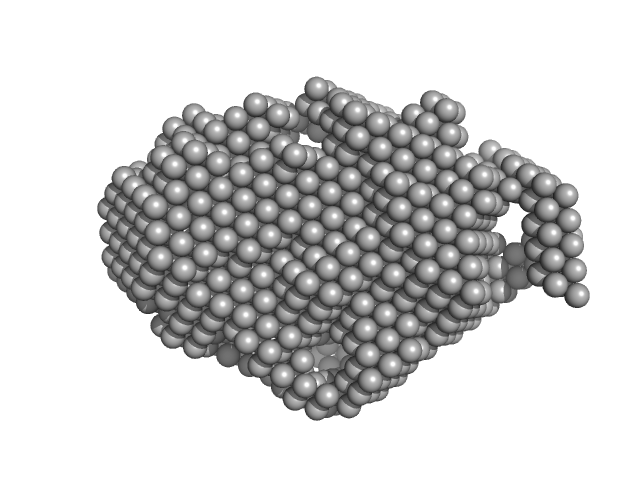
UniProt ID: F2L613 (1-401) Trehalose transferase (Trehalose phosphorylase/synthase)
|
|
|
|
| Sample: |
Trehalose transferase (Trehalose phosphorylase/synthase) monomer, 47 kDa Thermoproteus uzoniensis protein
|
| Buffer: |
50 mM HEPES, 100 NaCl, 4 mM DTT,, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 27
|
Anomeric Selectivity of Trehalose Transferase with Rare L-Sugars
ACS Catalysis (2020)
Mestrom L, Marsden S, van der Eijk H, Laustsen J, Jeffries C, Svergun D, Hagedoorn P, Bento I, Hanefeld U
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
UniProt ID: F2L613 (1-401) Trehalose transferase (Trehalose phosphorylase/synthase)
|
|
|

|
| Sample: |
Trehalose transferase (Trehalose phosphorylase/synthase) monomer, 47 kDa Thermoproteus uzoniensis protein
|
| Buffer: |
50 mM HEPES, 100 NaCl, 4 mM DTT, 20 mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 27
|
Anomeric Selectivity of Trehalose Transferase with Rare L-Sugars
ACS Catalysis (2020)
Mestrom L, Marsden S, van der Eijk H, Laustsen J, Jeffries C, Svergun D, Hagedoorn P, Bento I, Hanefeld U
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
UniProt ID: F2L613 (1-401) Trehalose transferase (Trehalose phosphorylase/synthase)
|
|
|

|
| Sample: |
Trehalose transferase (Trehalose phosphorylase/synthase) monomer, 47 kDa Thermoproteus uzoniensis protein
|
| Buffer: |
50 mM HEPES, 100 NaCl, 4 mM DTT, 20 mM MgCl2, 1 mM trehalose, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 27
|
Anomeric Selectivity of Trehalose Transferase with Rare L-Sugars
ACS Catalysis (2020)
Mestrom L, Marsden S, van der Eijk H, Laustsen J, Jeffries C, Svergun D, Hagedoorn P, Bento I, Hanefeld U
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
76 |
nm3 |
|
|