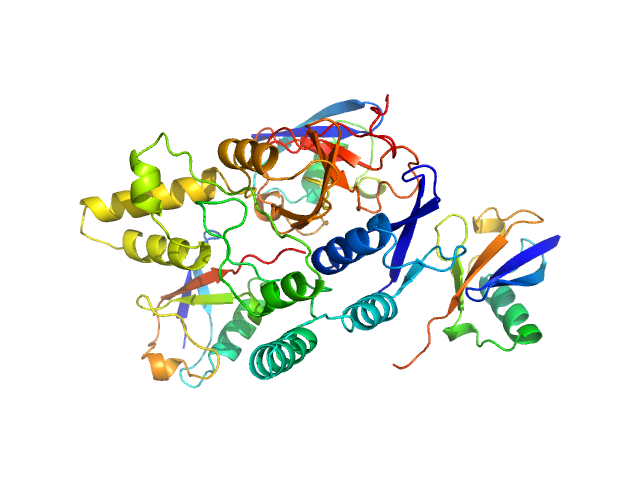
UniProt ID: Q8NBR6 (241-504) Ubiquitin carboxyl-terminal hydrolase C266A mutant
UniProt ID: P0CG48 (1-76) Polyubiquitin-C
|
|
|
|
| Sample: |
Ubiquitin carboxyl-terminal hydrolase C266A mutant monomer, 31 kDa Homo sapiens protein
Polyubiquitin-C trimer, 26 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 26
|
Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2.
Mol Cell (2021)
Abdul Rehman SA, Armstrong LA, Lange SM, Kristariyanto YA, Gräwert TW, Knebel A, Svergun DI, Kulathu Y
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
97 |
nm3 |
|
|
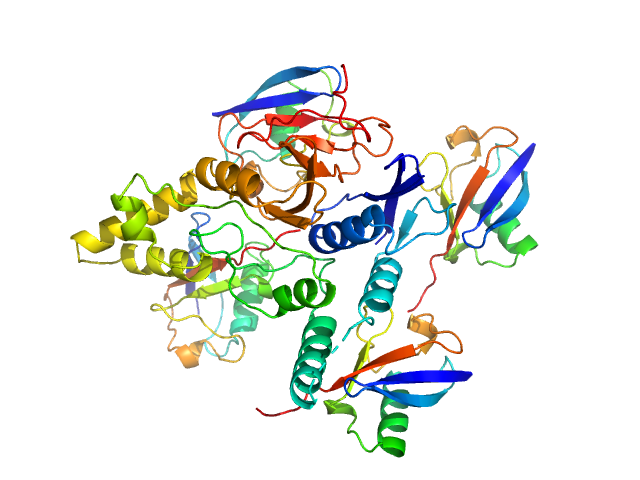
UniProt ID: Q8NBR6 (241-504) Ubiquitin carboxyl-terminal hydrolase C266A mutant
UniProt ID: P0CG48 (1-76) Polyubiquitin-C
|
|
|
|
| Sample: |
Ubiquitin carboxyl-terminal hydrolase C266A mutant monomer, 31 kDa Homo sapiens protein
Polyubiquitin-C tetramer, 34 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 26
|
Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2.
Mol Cell (2021)
Abdul Rehman SA, Armstrong LA, Lange SM, Kristariyanto YA, Gräwert TW, Knebel A, Svergun DI, Kulathu Y
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
111 |
nm3 |
|
|
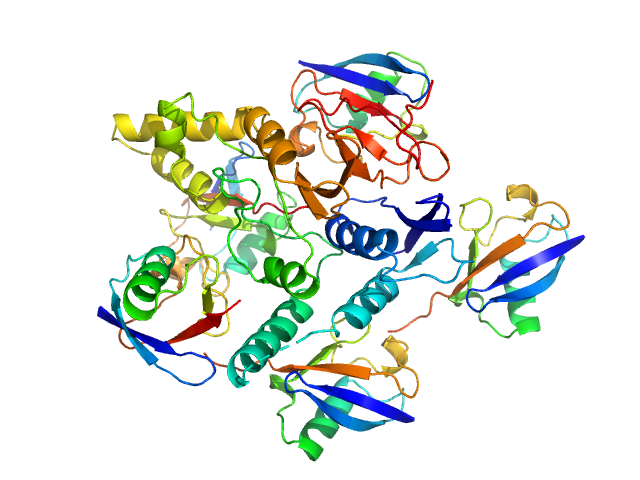
UniProt ID: Q8NBR6 (241-504) Ubiquitin carboxyl-terminal hydrolase C266A mutant
UniProt ID: P0CG48 (1-76) Polyubiquitin-C
|
|
|
|
| Sample: |
Ubiquitin carboxyl-terminal hydrolase C266A mutant monomer, 31 kDa Homo sapiens protein
Polyubiquitin-C pentamer, 43 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 26
|
Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2.
Mol Cell (2021)
Abdul Rehman SA, Armstrong LA, Lange SM, Kristariyanto YA, Gräwert TW, Knebel A, Svergun DI, Kulathu Y
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
116 |
nm3 |
|
|
UniProt ID: P15121 (1-316) Aldo-keto reductase family 1 member B1
|
|
|
|
| Sample: |
Aldo-keto reductase family 1 member B1 monomer, 36 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 250 mM NaCl, 100 mM 5-methyl uridine, pH: 8 |
| Experiment: |
SAXS
data collected at BL11 - NCD, ALBA on 2019 Jun 14
|
Improving data quality and expanding BioSAXS experiments to low-molecular-weight and low-concentration protein samples.
Acta Crystallogr D Struct Biol 76(Pt 10):971-981 (2020)
Castellví A, Pascual-Izarra C, Crosas E, Malfois M, Juanhuix J
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
UniProt ID: P00432 (1-527) Catalase
|
|
|
|
| Sample: |
Catalase tetramer, 240 kDa Bos taurus protein
|
| Buffer: |
50 mM sodium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at BL11 - NCD, ALBA on 2019 Jun 14
|
Improving data quality and expanding BioSAXS experiments to low-molecular-weight and low-concentration protein samples.
Acta Crystallogr D Struct Biol 76(Pt 10):971-981 (2020)
Castellví A, Pascual-Izarra C, Crosas E, Malfois M, Juanhuix J
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
265 |
nm3 |
|
|
UniProt ID: P00698 (1-147) Lysozyme C
|
|
|
|
| Sample: |
Lysozyme C monomer, 16 kDa Gallus gallus protein
|
| Buffer: |
40 mM sodium acetate, 150 mM NaCl, 100 mM 5-methyl uridine, pH: 3.8 |
| Experiment: |
SAXS
data collected at BL11 - NCD, ALBA on 2019 Jun 7
|
Improving data quality and expanding BioSAXS experiments to low-molecular-weight and low-concentration protein samples.
Acta Crystallogr D Struct Biol 76(Pt 10):971-981 (2020)
Castellví A, Pascual-Izarra C, Crosas E, Malfois M, Juanhuix J
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.2 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
UniProt ID: P07751 (965-1025) Spectrin alpha chain, non-erythrocytic 1
|
|
|
|
| Sample: |
Spectrin alpha chain, non-erythrocytic 1, 7 kDa Gallus gallus protein
|
| Buffer: |
10 mM citric acid, 150 mM NaCl, 100 mM 5-methyl uridine, pH: 3 |
| Experiment: |
SAXS
data collected at BL11 - NCD, ALBA on 2019 Jun 14
|
Improving data quality and expanding BioSAXS experiments to low-molecular-weight and low-concentration protein samples.
Acta Crystallogr D Struct Biol 76(Pt 10):971-981 (2020)
Castellví A, Pascual-Izarra C, Crosas E, Malfois M, Juanhuix J
|
| RgGuinier |
1.3 |
nm |
| Dmax |
4.5 |
nm |
| VolumePorod |
10 |
nm3 |
|
|
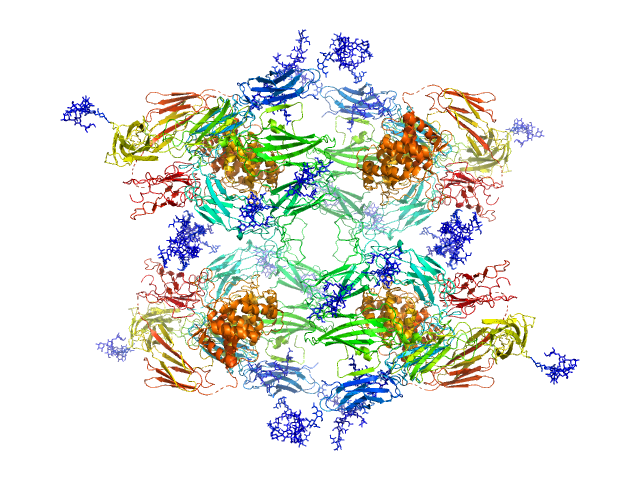
UniProt ID: P01023 (24-1474) Alpha-2-macroglobulin
|
|
|
|
| Sample: |
Alpha-2-macroglobulin tetramer, 643 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Bruker Nanostar w Excillum source, Department of Chemistry, iNANO building, Aarhus Uinversity on 2020 Jan 22
|
Structural Investigations of Human A2M Identify a Hollow Native Conformation That Underlies Its Distinctive Protease-Trapping Mechanism.
Mol Cell Proteomics 20:100090 (2021)
Harwood SL, Lyngsø J, Zarantonello A, Kjøge K, Nielsen PK, Andersen GR, Pedersen JS, Enghild JJ
|
| RgGuinier |
7.7 |
nm |
| Dmax |
22.7 |
nm |
|
|
UniProt ID: P01023 (24-1474) Alpha-2-macroglobulin
|
|
|
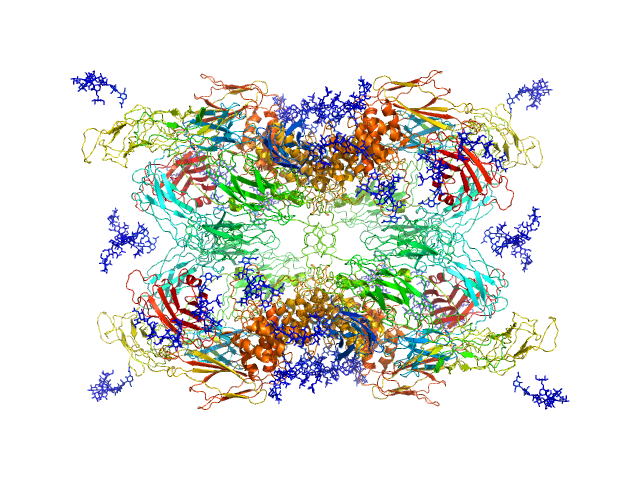
|
| Sample: |
Alpha-2-macroglobulin tetramer, 643 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Bruker Nanostar w Excillum source, Department of Chemistry, iNANO building, Aarhus Uinversity on 2020 Jan 22
|
Structural Investigations of Human A2M Identify a Hollow Native Conformation That Underlies Its Distinctive Protease-Trapping Mechanism.
Mol Cell Proteomics 20:100090 (2021)
Harwood SL, Lyngsø J, Zarantonello A, Kjøge K, Nielsen PK, Andersen GR, Pedersen JS, Enghild JJ
|
| RgGuinier |
6.7 |
nm |
| Dmax |
19.7 |
nm |
|
|
UniProt ID: P00760 (1-246) Cationic trypsin
UniProt ID: P01023 (24-1474) Alpha-2-macroglobulin
|
|
|
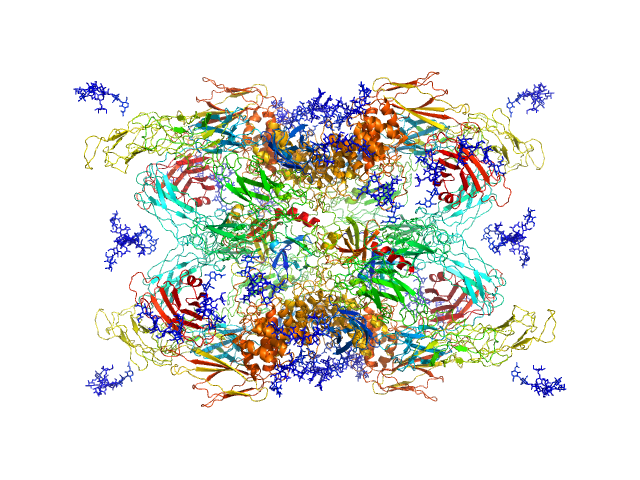
|
| Sample: |
Cationic trypsin dimer, 52 kDa Bos taurus protein
Alpha-2-macroglobulin tetramer, 643 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Bruker Nanostar w Excillum source, Department of Chemistry, iNANO building, Aarhus Uinversity on 2020 Jan 15
|
Structural Investigations of Human A2M Identify a Hollow Native Conformation That Underlies Its Distinctive Protease-Trapping Mechanism.
Mol Cell Proteomics 20:100090 (2021)
Harwood SL, Lyngsø J, Zarantonello A, Kjøge K, Nielsen PK, Andersen GR, Pedersen JS, Enghild JJ
|
| RgGuinier |
6.6 |
nm |
| Dmax |
19.7 |
nm |
|
|