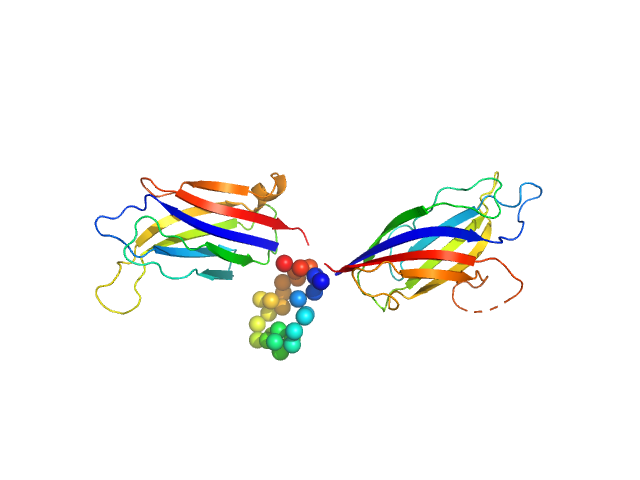
UniProt ID: Q9SKR2 (253-541) Synaptotagmin-1
|
|
|
|
| Sample: |
Synaptotagmin-1 monomer, 33 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Jun 11
|
The structure and flexibility analysis of the Arabidopsis
synaptotagmin 1 reveal the basis of its regulation at membrane contact sites
Life Science Alliance 4(10):e202101152 (2021)
Benavente J, Siliqi D, Infantes L, Lagartera L, Mills A, Gago F, Ruiz-López N, Botella M, Sánchez-Barrena M, Albert A
|
| RgGuinier |
3.1 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
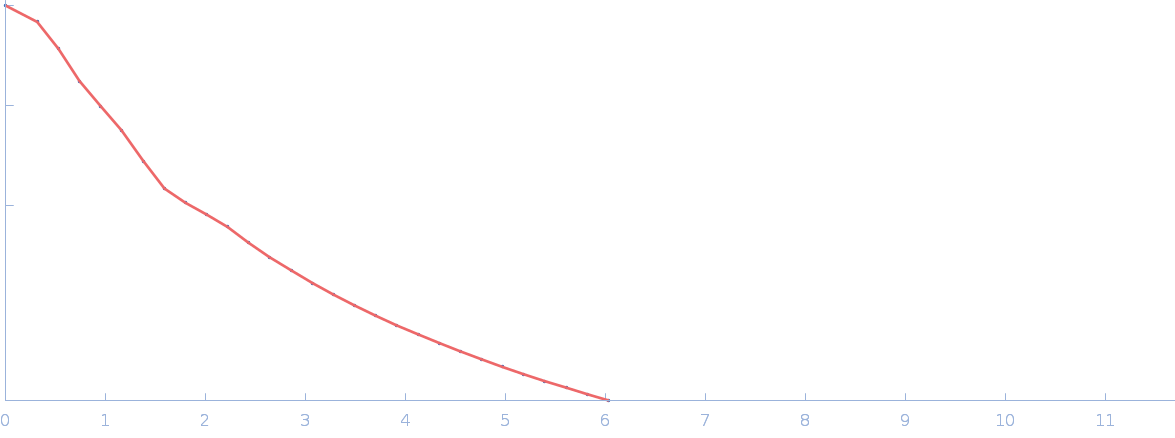
UniProt ID: P17846 (1-570) Sulfite reductase [NADPH] hemoprotein beta-component (Assimilatory NADPH-dependent sulfite reductase hemoprotein)
UniProt ID: P38038 (226-374) Sulfite reductase [NADPH] flavoprotein alpha-component (Assimilatory NADPH-dependent sulfite reductase flavoprotein)
|
|
|
|
| Sample: |
Sulfite reductase [NADPH] hemoprotein beta-component (Assimilatory NADPH-dependent sulfite reductase hemoprotein) monomer, 64 kDa Escherichia coli (strain … protein
Sulfite reductase [NADPH] flavoprotein alpha-component (Assimilatory NADPH-dependent sulfite reductase flavoprotein) monomer, 42 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM KPi, 100 mM NaCl, 1 mM EDTA, pH: 7.8 |
| Experiment: |
SANS
data collected at EQ-SANS (BL-6), Spallation Neutron Source on 2018 Jul 11
|
Small-angle neutron scattering solution structures of NADPH-dependent sulfite reductase
Journal of Structural Biology 213(2):107724 (2021)
Murray D, Weiss K, Stanley C, Nagy G, Stroupe M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.9 |
nm |
|
|
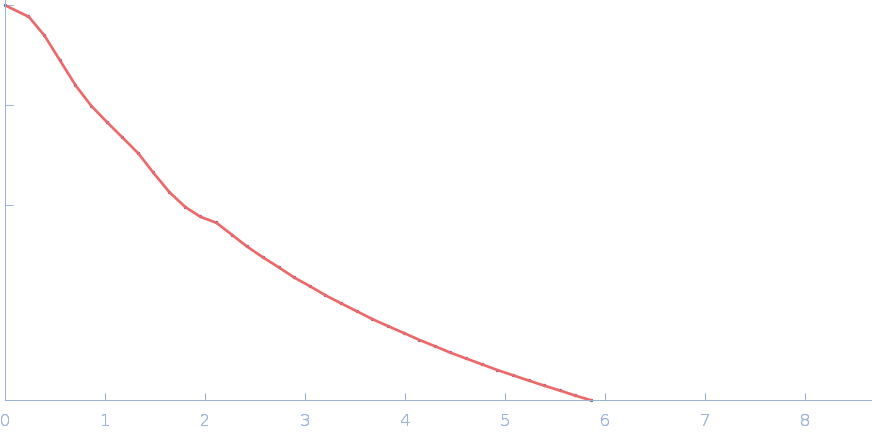
UniProt ID: P17846 (1-570) Sulfite reductase [NADPH] hemoprotein beta-component (Assimilatory NADPH-dependent sulfite reductase hemoprotein)
UniProt ID: P38038 (53-599) Sulfite reductase [NADPH] flavoprotein alpha-component (Assimilatory NADPH-dependent sulfite reductase flavoprotein)
|
|
|
|
| Sample: |
Sulfite reductase [NADPH] hemoprotein beta-component (Assimilatory NADPH-dependent sulfite reductase hemoprotein) monomer, 64 kDa Escherichia coli (strain … protein
Sulfite reductase [NADPH] flavoprotein alpha-component (Assimilatory NADPH-dependent sulfite reductase flavoprotein) monomer, 61 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM KPi, 100 mM NaCl, 1 mM EDTA, pH: 7.8 |
| Experiment: |
SANS
data collected at EQ-SANS (BL-6), Spallation Neutron Source on 2018 Jul 11
|
Small-angle neutron scattering solution structures of NADPH-dependent sulfite reductase
Journal of Structural Biology 213(2):107724 (2021)
Murray D, Weiss K, Stanley C, Nagy G, Stroupe M
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.4 |
nm |
|
|
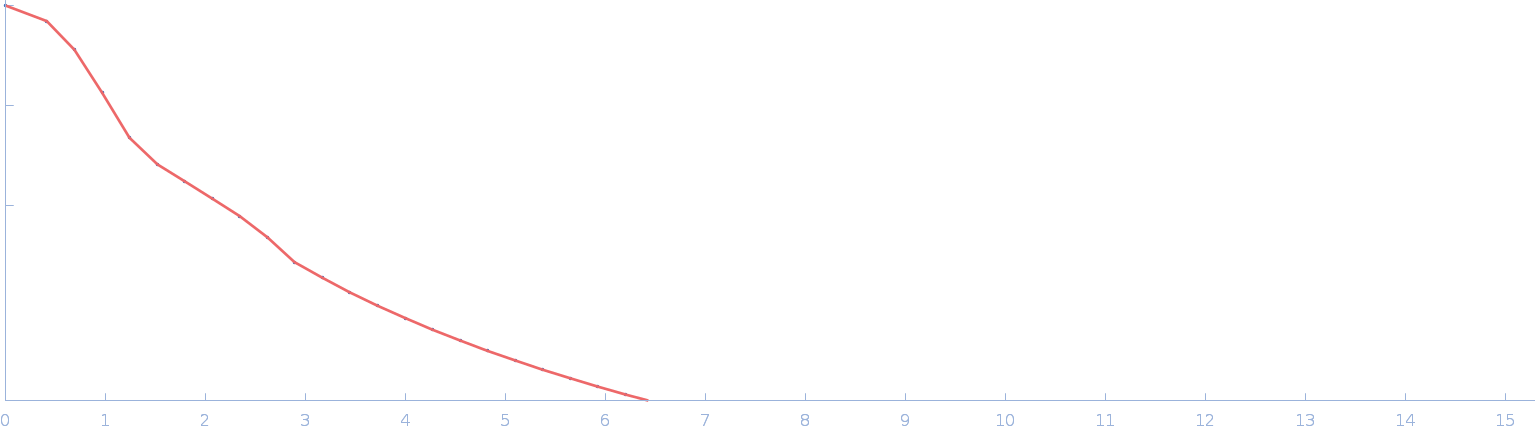
UniProt ID: P17846 (1-570) Sulfite reductase [NADPH] hemoprotein beta-component (Assimilatory NADPH-dependent sulfite reductase hemoprotein)
|
|
|
|
| Sample: |
Sulfite reductase [NADPH] hemoprotein beta-component (Assimilatory NADPH-dependent sulfite reductase hemoprotein) monomer, 64 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM KPi, 100 mM NaCl, 1 mM EDTA, pH: 7.8 |
| Experiment: |
SANS
data collected at EQ-SANS (BL-6), Spallation Neutron Source on 2020 May 28
|
Small-angle neutron scattering solution structures of NADPH-dependent sulfite reductase
Journal of Structural Biology 213(2):107724 (2021)
Murray D, Weiss K, Stanley C, Nagy G, Stroupe M
|
| RgGuinier |
2.6 |
nm |
| Dmax |
7.5 |
nm |
|
|
UniProt ID: P38038 (53-599) Sulfite reductase [NADPH] flavoprotein alpha-component (Assimilatory NADPH-dependent sulfite reductase flavoprotein)
|
|
|
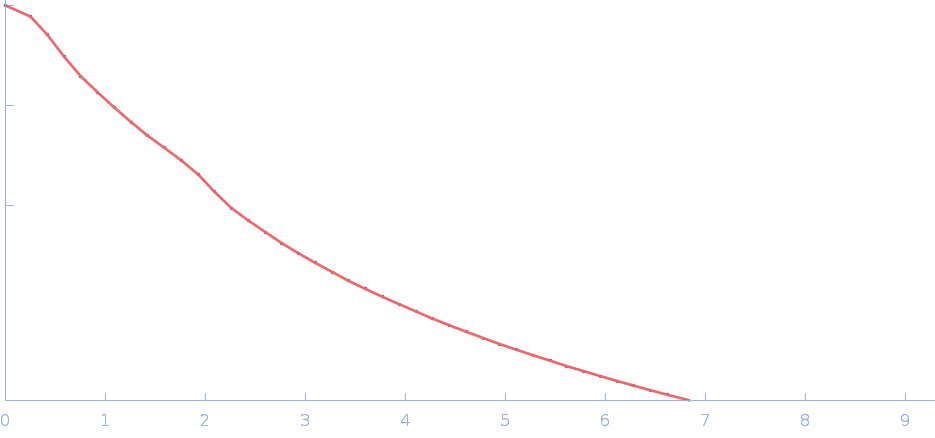
|
| Sample: |
Sulfite reductase [NADPH] flavoprotein alpha-component (Assimilatory NADPH-dependent sulfite reductase flavoprotein) monomer, 61 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM KPi, 100 mM NaCl, 1 mM EDTA, pH: 7.8 |
| Experiment: |
SANS
data collected at EQ-SANS (BL-6), Spallation Neutron Source on 2018 Jul 11
|
Small-angle neutron scattering solution structures of NADPH-dependent sulfite reductase
Journal of Structural Biology 213(2):107724 (2021)
Murray D, Weiss K, Stanley C, Nagy G, Stroupe M
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.2 |
nm |
|
|
UniProt ID: P17846 (1-570) Sulfite reductase [NADPH] hemoprotein beta-component (Assimilatory NADPH-dependent sulfite reductase hemoprotein)
|
|
|
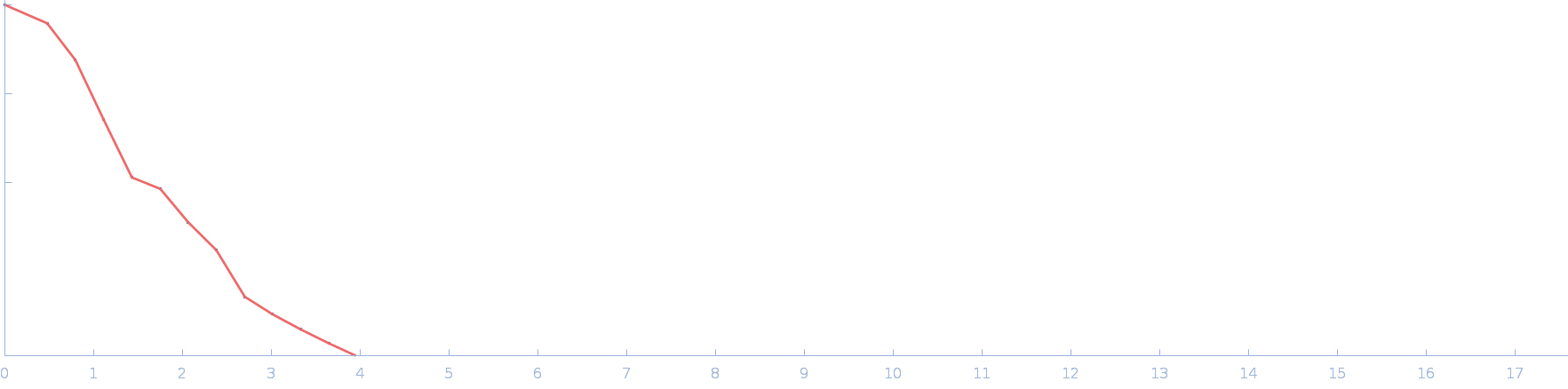
|
| Sample: |
Sulfite reductase [NADPH] hemoprotein beta-component (Assimilatory NADPH-dependent sulfite reductase hemoprotein) monomer, 64 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM KPi, 100 mM NaCl, 1 mM EDTA, pH: 7.8 |
| Experiment: |
SANS
data collected at EQ-SANS (BL-6), Spallation Neutron Source on 2020 Jun 28
|
Small-angle neutron scattering solution structures of NADPH-dependent sulfite reductase
Journal of Structural Biology 213(2):107724 (2021)
Murray D, Weiss K, Stanley C, Nagy G, Stroupe M
|
| RgGuinier |
2.4 |
nm |
| Dmax |
6.6 |
nm |
|
|
UniProt ID: P38038 (53-599) Sulfite reductase [NADPH] flavoprotein alpha-component (Assimilatory NADPH-dependent sulfite reductase flavoprotein)
|
|
|
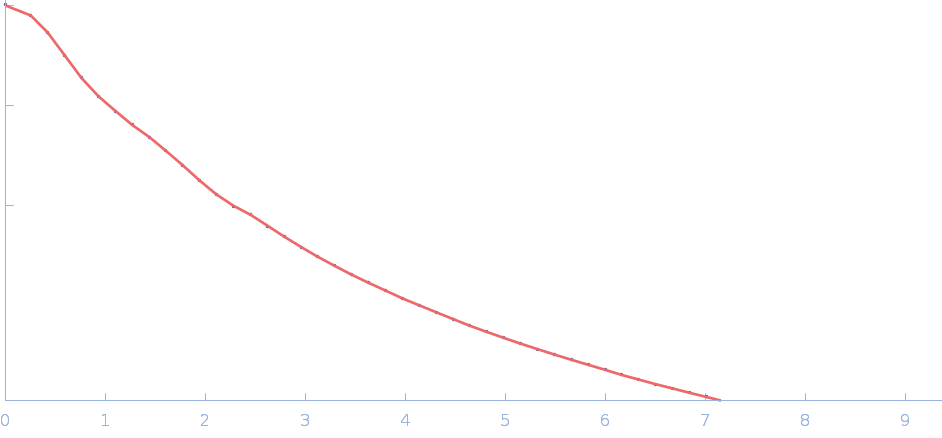
|
| Sample: |
Sulfite reductase [NADPH] flavoprotein alpha-component (Assimilatory NADPH-dependent sulfite reductase flavoprotein) monomer, 61 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM KPi, 100 mM NaCl, 1 mM EDTA, pH: 7.8 |
| Experiment: |
SANS
data collected at EQ-SANS (BL-6), Spallation Neutron Source on 2020 Jun 26
|
Small-angle neutron scattering solution structures of NADPH-dependent sulfite reductase
Journal of Structural Biology 213(2):107724 (2021)
Murray D, Weiss K, Stanley C, Nagy G, Stroupe M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
12.4 |
nm |
|
|
UniProt ID: P0DTD1 (5926-6452) Replicase polyprotein 1ab (non-structural protein 14)
|
|
|
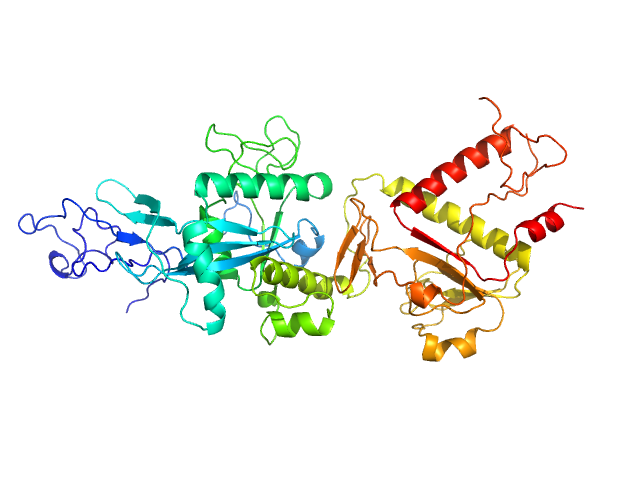
|
| Sample: |
Replicase polyprotein 1ab (non-structural protein 14) monomer, 60 kDa Severe acute respiratory … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 5 mM MgCl2, 2 mM β-mercaptoethanol, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Dec 11
|
Despite the odds: formation of the SARS-CoV-2 methylation complex
Nucleic Acids Research (2024)
Matsuda A, Plewka J, Rawski M, Mourão A, Zajko W, Siebenmorgen T, Kresik L, Lis K, Jones A, Pachota M, Karim A, Hartman K, Nirwal S, Sonani R, Chykunova Y, Minia I, Mak P, Landthaler M, Nowotny M, Dubin G, Sattler M, Suder P, Popowicz G, Pyrć K, Czarna A
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
80 |
nm3 |
|
|
UniProt ID: P0DTD1 (5926-6452) Replicase polyprotein 1ab (non-structural protein 14)
UniProt ID: P0DTC1 (4254-4392) Replicase polyprotein 1a (non-structural protein 10)
|
|
|
|
| Sample: |
Replicase polyprotein 1ab (non-structural protein 14) monomer, 60 kDa Severe acute respiratory … protein
Replicase polyprotein 1a (non-structural protein 10) monomer, 15 kDa Severe acute respiratory … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 5 mM MgCl2, 2 mM β-mercaptoethanol, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Dec 11
|
Despite the odds: formation of the SARS-CoV-2 methylation complex
Nucleic Acids Research (2024)
Matsuda A, Plewka J, Rawski M, Mourão A, Zajko W, Siebenmorgen T, Kresik L, Lis K, Jones A, Pachota M, Karim A, Hartman K, Nirwal S, Sonani R, Chykunova Y, Minia I, Mak P, Landthaler M, Nowotny M, Dubin G, Sattler M, Suder P, Popowicz G, Pyrć K, Czarna A
|
| RgGuinier |
2.9 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
73 |
nm3 |
|
|
UniProt ID: P0DTC1 (4254-4392) Replicase polyprotein 1a (non-structural protein 10)
UniProt ID: P0DTD1 (6799-7096) Replicase polyprotein 1ab (non-structural protein 16)
|
|
|

|
| Sample: |
Replicase polyprotein 1a (non-structural protein 10) monomer, 15 kDa Severe acute respiratory … protein
Replicase polyprotein 1ab (non-structural protein 16) monomer, 33 kDa Severe acute respiratory … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 5 mM MgCl2, 2 mM β-mercaptoethanol, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Dec 11
|
Despite the odds: formation of the SARS-CoV-2 methylation complex
Nucleic Acids Research (2024)
Matsuda A, Plewka J, Rawski M, Mourão A, Zajko W, Siebenmorgen T, Kresik L, Lis K, Jones A, Pachota M, Karim A, Hartman K, Nirwal S, Sonani R, Chykunova Y, Minia I, Mak P, Landthaler M, Nowotny M, Dubin G, Sattler M, Suder P, Popowicz G, Pyrć K, Czarna A
|
| RgGuinier |
2.0 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
28 |
nm3 |
|
|