UniProt ID: P0CU08 (81-235) Survival of motor neuron protein-interacting protein yip11 (Gemin2 ΔN80)
UniProt ID: Q09808 (None-None) Survival motor neuron-like protein 1 (Δ36-119)
|
|
|
|
| Sample: |
Survival of motor neuron protein-interacting protein yip11 (Gemin2 ΔN80) monomer, 18 kDa Schizosaccharomyces pombe (strain … protein
Survival motor neuron-like protein 1 (Δ36-119) monomer, 8 kDa Schizosaccharomyces pombe protein
|
| Buffer: |
150 mM NaCl, 20 mM HEPES, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Apr 30
|
Identification and structural analysis of the Schizosaccharomyces pombe
SMN complex
Nucleic Acids Research (2021)
Veepaschit J, Viswanathan A, Bordonné R, Grimm C, Fischer U
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.0 |
nm |
|
|
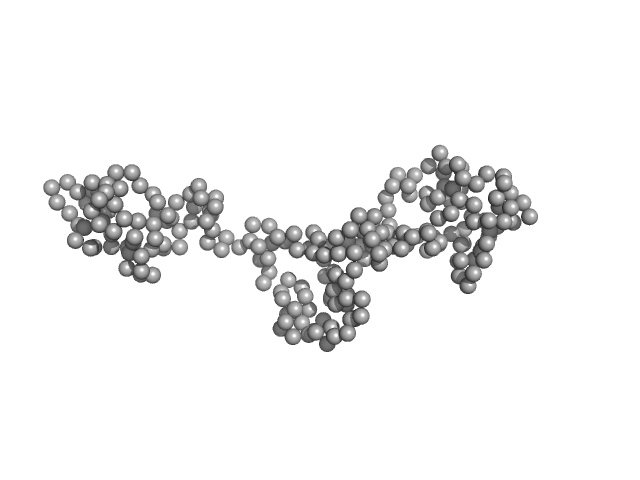
UniProt ID: Q57W63 (476-523) Coronin
|
|
|
|
| Sample: |
Coronin tetramer, 21 kDa Trypanosoma brucei brucei … protein
|
| Buffer: |
100 mM Tris-HCl, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2019 Oct 17
|
Structural insights into kinetoplastid coronin oligomerization domain and F-actin interaction
Current Research in Structural Biology (2021)
Parihar P, Singh A, Karade S, Sahasrabuddhe A, Pratap J
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
UniProt ID: E9B8V9 (1-223) Cell cycle associated protein MOB1, putative
|
|
|
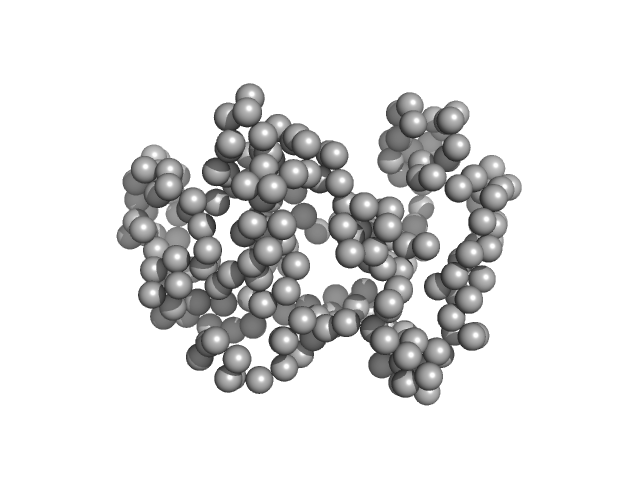
|
| Sample: |
Cell cycle associated protein MOB1, putative monomer, 27 kDa Leishmania donovani (strain … protein
|
| Buffer: |
100 mM Tris-HCl, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2017 Jun 22
|
Structural insights into kinetoplastid coronin oligomerization domain and F-actin interaction
Current Research in Structural Biology (2021)
Parihar P, Singh A, Karade S, Sahasrabuddhe A, Pratap J
|
| RgGuinier |
2.1 |
nm |
| Dmax |
5.6 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
UniProt ID: P0CU08 (81-235) Survival of motor neuron protein-interacting protein yip11 (Gemin2 ΔN80)
UniProt ID: Q09808 (None-None) Survival motor neuron-like protein 1 (Δ36-119)
|
|
|
|
| Sample: |
Survival of motor neuron protein-interacting protein yip11 (Gemin2 ΔN80) monomer, 18 kDa Schizosaccharomyces pombe (strain … protein
Survival motor neuron-like protein 1 (Δ36-119) monomer, 8 kDa Schizosaccharomyces pombe protein
|
| Buffer: |
150 mM NaCl, 20 mM HEPES, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Apr 29
|
Identification and structural analysis of the Schizosaccharomyces pombe
SMN complex
Nucleic Acids Research (2021)
Veepaschit J, Viswanathan A, Bordonné R, Grimm C, Fischer U
|
| RgGuinier |
4.8 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
386 |
nm3 |
|
|
UniProt ID: Q9Y2S7 (51-368) Polymerase delta-interacting protein 2
|
|
|
|
| Sample: |
Polymerase delta-interacting protein 2 monomer, 37 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES pH 7.5, 500 mM NaCl, 5% (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar, University of Huddersfield on 2019 Jul 18
|
Crystal structure and molecular dynamics of human POLDIP2, a multifaceted adaptor protein in metabolism and genome stability.
Protein Sci (2021)
Kulik AA, Maruszczak KK, Thomas DC, Nabi-Aldridge NLA, Carr M, Bingham RJ, Cooper CDO
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
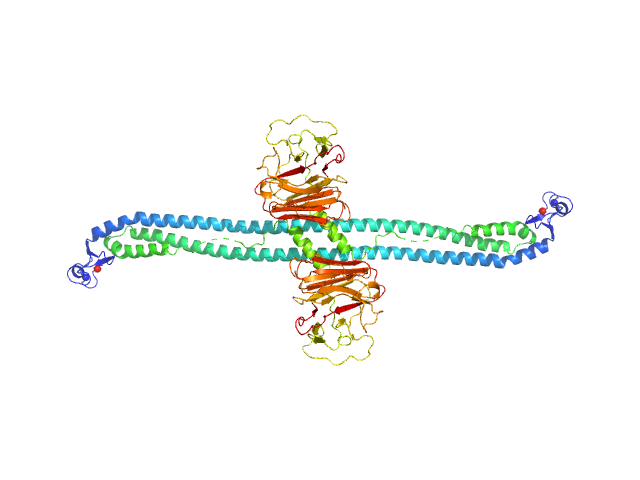
UniProt ID: Q1XH17 (2-477) Tripartite motif-containing protein 72
|
|
|

|
| Sample: |
Tripartite motif-containing protein 72 dimer, 105 kDa Mus musculus protein
|
| Buffer: |
25 mM Tris-HCl, pH 8.0, 300 mM NaCl, 1 mM TCEP, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2019 Dec 7
|
Structure and activation of the RING E3 ubiquitin ligase TRIM72 on the membrane.
Nat Struct Mol Biol (2023)
Park SH, Han J, Jeong BC, Song JH, Jang SH, Jeong H, Kim BH, Ko YG, Park ZY, Lee KE, Hyun J, Song HK
|
| RgGuinier |
6.9 |
nm |
| Dmax |
24.0 |
nm |
| VolumePorod |
384 |
nm3 |
|
|
UniProt ID: Q1XH17 (79-470) Tripartite motif-containing protein 72
|
|
|
|
| Sample: |
Tripartite motif-containing protein 72 dimer, 88 kDa Mus musculus protein
|
| Buffer: |
25 mM Tris-HCl, pH 8.0, 300 mM NaCl, 1 mM TCEP, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2019 Dec 7
|
Structure and activation of the RING E3 ubiquitin ligase TRIM72 on the membrane.
Nat Struct Mol Biol (2023)
Park SH, Han J, Jeong BC, Song JH, Jang SH, Jeong H, Kim BH, Ko YG, Park ZY, Lee KE, Hyun J, Song HK
|
| RgGuinier |
5.1 |
nm |
| Dmax |
18.7 |
nm |
| VolumePorod |
142 |
nm3 |
|
|
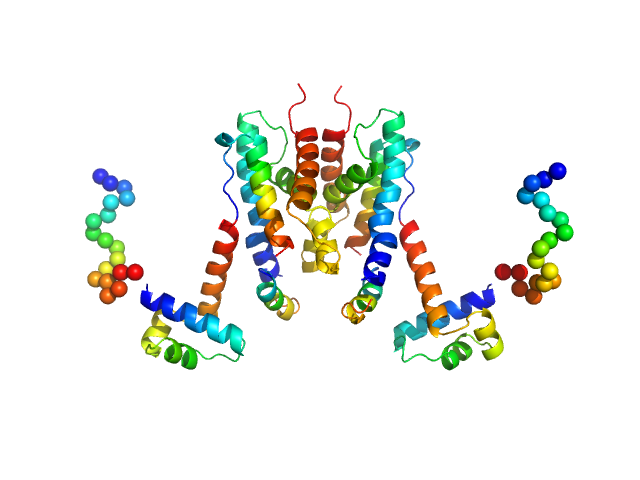
UniProt ID: A0A2A6N3G4 (19-229) TetR/AcrR family transcriptional regulator
|
|
|
|
| Sample: |
TetR/AcrR family transcriptional regulator dimer, 48 kDa Bradyrhizobium diazoefficiens protein
|
| Buffer: |
20 mM Na2HPO4, 50 mM NaCl, 50 mM imidazole, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Sep 23
|
The Induction Mechanism of the Flavonoid-Responsive Regulator FrrA.
FEBS J (2021)
Werner N, Werten S, Hoppen J, Palm GJ, Göttfert M, Hinrichs W
|
| RgGuinier |
3.4 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
93 |
nm3 |
|
|
UniProt ID: A0A2A6N3G4 (19-229) TetR/AcrR family transcriptional regulator
UniProt ID: None (None-None) 5,7-dihydroxy-3-(4-hydroxyphenyl)chromen-4-one
|
|
|
|
| Sample: |
TetR/AcrR family transcriptional regulator dimer, 48 kDa Bradyrhizobium diazoefficiens protein
5,7-dihydroxy-3-(4-hydroxyphenyl)chromen-4-one monomer, 0 kDa
|
| Buffer: |
20 mM Na2HPO4, 50 mM NaCl, 50 mM imidazole, 0.08 mM genistein, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Oct 27
|
The Induction Mechanism of the Flavonoid-Responsive Regulator FrrA.
FEBS J (2021)
Werner N, Werten S, Hoppen J, Palm GJ, Göttfert M, Hinrichs W
|
| RgGuinier |
3.4 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
96 |
nm3 |
|
|
UniProt ID: A0A2A6N3G4 (19-229) TetR/AcrR family transcriptional regulator
UniProt ID: None (None-None) 5,7-dihydroxy-2-(4-hydroxyphenyl)chroman-4-one
|
|
|
|
| Sample: |
TetR/AcrR family transcriptional regulator dimer, 48 kDa Bradyrhizobium diazoefficiens protein
5,7-dihydroxy-2-(4-hydroxyphenyl)chroman-4-one monomer, 0 kDa
|
| Buffer: |
20 mM Na2HPO4, 50 mM NaCl, 50 mM imidazole, 1 mM naringenin, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Sep 23
|
The Induction Mechanism of the Flavonoid-Responsive Regulator FrrA.
FEBS J (2021)
Werner N, Werten S, Hoppen J, Palm GJ, Göttfert M, Hinrichs W
|
| RgGuinier |
3.2 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
91 |
nm3 |
|
|