UniProt ID: P0DTD1 (5926-6452) Replicase polyprotein 1ab (non-structural protein 14)
UniProt ID: P0DTC1 (4254-4392) Replicase polyprotein 1a (non-structural protein 10)
UniProt ID: P0DTD1 (6799-7096) Replicase polyprotein 1ab (non-structural protein 16)
|
|
|

|
| Sample: |
Replicase polyprotein 1ab (non-structural protein 14) monomer, 60 kDa Severe acute respiratory … protein
Replicase polyprotein 1a (non-structural protein 10) monomer, 15 kDa Severe acute respiratory … protein
Replicase polyprotein 1ab (non-structural protein 16) monomer, 33 kDa Severe acute respiratory … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 5 mM MgCl2, 2 mM β-mercaptoethanol, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Dec 11
|
Despite the odds: formation of the SARS-CoV-2 methylation complex
Nucleic Acids Research (2024)
Matsuda A, Plewka J, Rawski M, Mourão A, Zajko W, Siebenmorgen T, Kresik L, Lis K, Jones A, Pachota M, Karim A, Hartman K, Nirwal S, Sonani R, Chykunova Y, Minia I, Mak P, Landthaler M, Nowotny M, Dubin G, Sattler M, Suder P, Popowicz G, Pyrć K, Czarna A
|
| RgGuinier |
4.7 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
113 |
nm3 |
|
|
UniProt ID: P58093 (1-80) Antitoxin ParD
|
|
|
|
| Sample: |
Antitoxin ParD dodecamer, 108 kDa Vibrio cholerae serotype … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Jul 18
|
Entropic pressure controls the oligomerization of the Vibrio cholerae
ParD2 antitoxin
Acta Crystallographica Section D Structural Biology 77(7):904-920 (2021)
Garcia-Rodriguez G, Girardin Y, Volkov A, Singh R, Muruganandam G, Van Dyck J, Sobott F, Versées W, Charlier D, Loris R
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
190 |
nm3 |
|
|
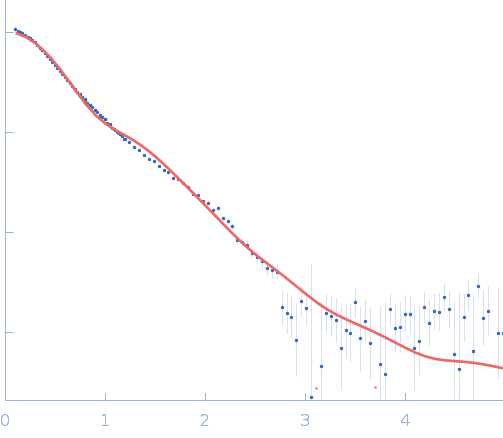
UniProt ID: P38038 (53-599) Sulfite reductase [NADPH] flavoprotein alpha-component (Assimilatory NADPH-dependent sulfite reductase flavoprotein)
|
|
|
|
| Sample: |
Sulfite reductase [NADPH] flavoprotein alpha-component (Assimilatory NADPH-dependent sulfite reductase flavoprotein) monomer, 61 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM KPi, 100 mM NaCl, 1 mM EDTA, pH: 7.8 |
| Experiment: |
SANS
data collected at EQ-SANS (BL-6), Spallation Neutron Source on 2018 Jul 11
|
NADPH-dependent sulfite reductase flavoprotein adopts an extended conformation unique to this diflavin reductase
Journal of Structural Biology 205(2):170-179 (2019)
Tavolieri A, Murray D, Askenasy I, Pennington J, McGarry L, Stanley C, Stroupe M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
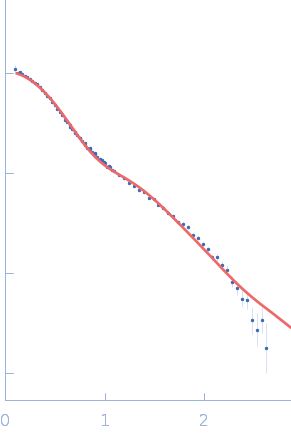
UniProt ID: P38038 (53-599) Sulfite reductase [NADPH] flavoprotein alpha-component (Assimilatory NADPH-dependent sulfite reductase flavoprotein)
|
|
|
|
| Sample: |
Sulfite reductase [NADPH] flavoprotein alpha-component (Assimilatory NADPH-dependent sulfite reductase flavoprotein) monomer, 61 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM KPi, 100 mM NaCl, 1 mM EDTA, pH: 7.8 |
| Experiment: |
SANS
data collected at EQ-SANS (BL-6), Spallation Neutron Source on 2018 Jul 11
|
NADPH-dependent sulfite reductase flavoprotein adopts an extended conformation unique to this diflavin reductase
Journal of Structural Biology 205(2):170-179 (2019)
Tavolieri A, Murray D, Askenasy I, Pennington J, McGarry L, Stanley C, Stroupe M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.3 |
nm |
| VolumePorod |
73 |
nm3 |
|
|
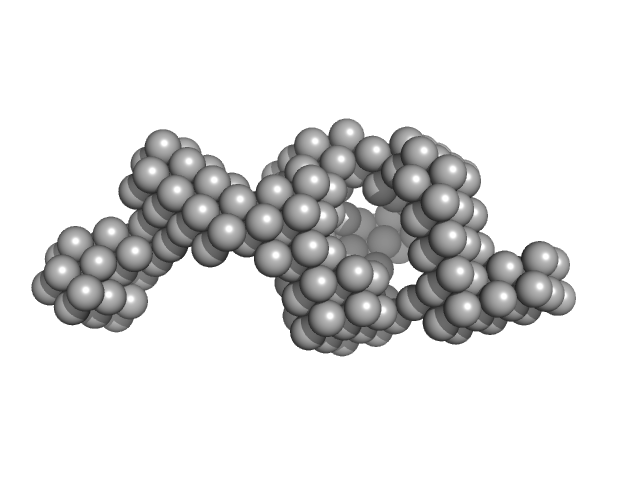
UniProt ID: P71888 (1-148) Probable transcriptional regulatory protein (Probably AsnC-family)
|
|
|
|
| Sample: |
Probable transcriptional regulatory protein (Probably AsnC-family) dimer, 36 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2019 Mar 1
|
Mycobacterium tuberculosis Rv2324 is a multifunctional feast/famine regulatory protein involved in growth, DNA replication and damage control.
Int J Biol Macromol 252:126459 (2023)
Dubey S, Maurya RK, Shree S, Kumar S, Jahan F, Krishnan MY, Ramachandran R
|
| RgGuinier |
2.7 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
156 |
nm3 |
|
|
UniProt ID: P71888 (1-148) Probable transcriptional regulatory protein (Probably AsnC-family)
|
|
|

|
| Sample: |
Probable transcriptional regulatory protein (Probably AsnC-family) octamer, 143 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM EDTA, 2 mM methionine, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2019 Dec 5
|
Mycobacterium tuberculosis Rv2324 is a multifunctional feast/famine regulatory protein involved in growth, DNA replication and damage control.
Int J Biol Macromol 252:126459 (2023)
Dubey S, Maurya RK, Shree S, Kumar S, Jahan F, Krishnan MY, Ramachandran R
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.8 |
nm |
| VolumePorod |
246 |
nm3 |
|
|
UniProt ID: Q8MQJ9 (756-1037) Brain tumor protein
UniProt ID: P25822 (1093-1426) Maternal protein pumilio
UniProt ID: P25724 (301-392) Protein nanos
UniProt ID: None (None-None) hunchback mRNA Nanos Response Element 2
|
|
|
|
| Sample: |
Brain tumor protein monomer, 32 kDa Drosophila melanogaster protein
Maternal protein pumilio monomer, 38 kDa Drosophila melanogaster protein
Protein nanos monomer, 11 kDa Drosophila melanogaster protein
Hunchback mRNA Nanos Response Element 2 monomer, 7 kDa Drosophila melanogaster RNA
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 1 mM DTT, 3% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Nov 12
|
Structure and dynamics of the quaternary hunchback mRNA translation repression complex.
Nucleic Acids Res 49(15):8866-8885 (2021)
Macošek J, Simon B, Linse JB, Jagtap PKA, Winter SL, Foot J, Lapouge K, Perez K, Rettel M, Ivanović MT, Masiewicz P, Murciano B, Savitski MM, Loedige I, Hub JS, Gabel F, Hennig J
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
114 |
nm3 |
|
|
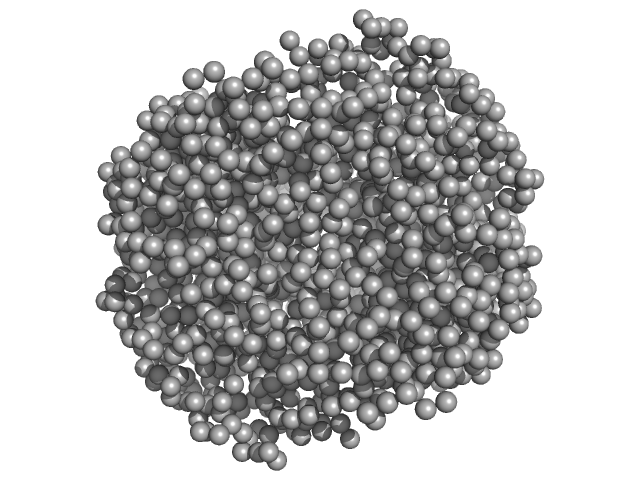
UniProt ID: None (None-None) HpcH/HpaI aldolase
|
|
|
|
| Sample: |
HpcH/HpaI aldolase hexamer, 165 kDa Rhizorhabdus wittichii RW1 protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 20 mg/l MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 15
|
Substrate Induced Movement of the Metal Cofactor between Active and Resting State
Angewandte Chemie International Edition (2022)
Marsden S, Wijma H, Mohr M, Justo I, Hagedoorn P, Laustsen J, Jeffries C, Svergun D, Mestrom L, McMillan D, Bento I, Hanefeld U
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
224 |
nm3 |
|
|
UniProt ID: None (None-None) HpcH/HpaI aldolase (S116A)
|
|
|
|
| Sample: |
HpcH/HpaI aldolase (S116A) hexamer, 165 kDa Sphingomonas wittichii (strain … protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 20 mg/l MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 15
|
Substrate Induced Movement of the Metal Cofactor between Active and Resting State
Angewandte Chemie International Edition (2022)
Marsden S, Wijma H, Mohr M, Justo I, Hagedoorn P, Laustsen J, Jeffries C, Svergun D, Mestrom L, McMillan D, Bento I, Hanefeld U
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
222 |
nm3 |
|
|
UniProt ID: None (None-None) HpcH/HpaI aldolase (S116C)
|
|
|
|
| Sample: |
HpcH/HpaI aldolase (S116C) hexamer, 165 kDa Rhizorhabdus wittichii RW1 protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 20 mg/l MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 15
|
Substrate Induced Movement of the Metal Cofactor between Active and Resting State
Angewandte Chemie International Edition (2022)
Marsden S, Wijma H, Mohr M, Justo I, Hagedoorn P, Laustsen J, Jeffries C, Svergun D, Mestrom L, McMillan D, Bento I, Hanefeld U
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
223 |
nm3 |
|
|