UniProt ID: Q13485 (10-140) Mothers against decapentaplegic homolog 4 (MH1 fragment)
|
|
|
|
| Sample: |
Mothers against decapentaplegic homolog 4 (MH1 fragment) monomer, 15 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Apr 20
|
Conformational landscape of multidomain SMAD proteins
Computational and Structural Biotechnology Journal (2021)
Gomes T, Martin-Malpartida P, Ruiz L, Aragón E, Cordeiro T, Macias M
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
UniProt ID: Q9SKR2 (253-541) Synaptotagmin-1
|
|
|
|
| Sample: |
Synaptotagmin-1 monomer, 33 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Feb 16
|
The structure and flexibility analysis of the Arabidopsis
synaptotagmin 1 reveal the basis of its regulation at membrane contact sites
Life Science Alliance 4(10):e202101152 (2021)
Benavente J, Siliqi D, Infantes L, Lagartera L, Mills A, Gago F, Ruiz-López N, Botella M, Sánchez-Barrena M, Albert A
|
| RgGuinier |
2.8 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
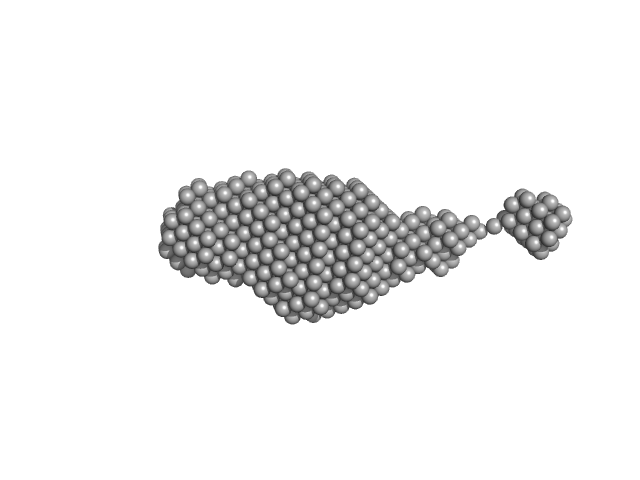
UniProt ID: Q9SKR2 (253-541) Synaptotagmin-1
|
|
|
|
| Sample: |
Synaptotagmin-1 monomer, 33 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Feb 16
|
The structure and flexibility analysis of the Arabidopsis
synaptotagmin 1 reveal the basis of its regulation at membrane contact sites
Life Science Alliance 4(10):e202101152 (2021)
Benavente J, Siliqi D, Infantes L, Lagartera L, Mills A, Gago F, Ruiz-López N, Botella M, Sánchez-Barrena M, Albert A
|
| RgGuinier |
2.9 |
nm |
| Dmax |
13.8 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
UniProt ID: B7UI23 (1-3223) Efa1/LifA protein
|
|
|

|
| Sample: |
Efa1/LifA protein monomer, 367 kDa Escherichia coli O127:H6 … protein
|
| Buffer: |
50 mM Bis-Tris, 100 mM NaCl, 3 % (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jun 29
|
Activity of lymphostatin, a lymphocyte inhibitory virulence factor of pathogenic Escherichia coli, is dependent on a cysteine protease motif.
J Mol Biol :167200 (2021)
Bease AG, Blackburn EA, Chintoan-Uta C, Webb S, Cassady-Cain RL, Stevens MP
|
| RgGuinier |
5.9 |
nm |
| Dmax |
19.3 |
nm |
| VolumePorod |
559 |
nm3 |
|
|
UniProt ID: B7UI23 (1-3223) Efa1/LifA protein
|
|
|
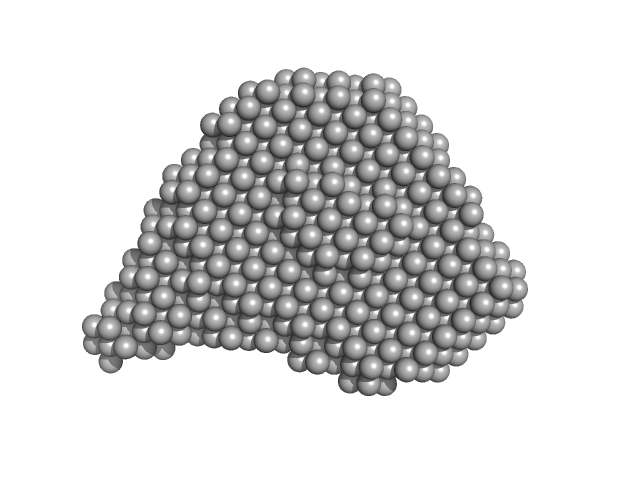
|
| Sample: |
Efa1/LifA protein monomer, 367 kDa Escherichia coli O127:H6 … protein
|
| Buffer: |
50 mM Bis-Tris, 100 mM NaCl, 3 % (v/v) glycerol, pH: 5.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jun 29
|
Activity of lymphostatin, a lymphocyte inhibitory virulence factor of pathogenic Escherichia coli, is dependent on a cysteine protease motif.
J Mol Biol :167200 (2021)
Bease AG, Blackburn EA, Chintoan-Uta C, Webb S, Cassady-Cain RL, Stevens MP
|
| RgGuinier |
5.9 |
nm |
| Dmax |
19.7 |
nm |
| VolumePorod |
565 |
nm3 |
|
|
UniProt ID: B7UI23 (1-3223) Efa1/LifA protein (C1480A mutant)
|
|
|

|
| Sample: |
Efa1/LifA protein (C1480A mutant) monomer, 367 kDa Escherichia coli O127:H6 … protein
|
| Buffer: |
50 mM Bis-Tris, 100 mM NaCl, 3 % (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jun 29
|
Activity of lymphostatin, a lymphocyte inhibitory virulence factor of pathogenic Escherichia coli, is dependent on a cysteine protease motif.
J Mol Biol :167200 (2021)
Bease AG, Blackburn EA, Chintoan-Uta C, Webb S, Cassady-Cain RL, Stevens MP
|
| RgGuinier |
5.9 |
nm |
| Dmax |
19.1 |
nm |
| VolumePorod |
557 |
nm3 |
|
|
UniProt ID: B7UI23 (1-3223) Efa1/LifA protein (C1480A mutant)
|
|
|
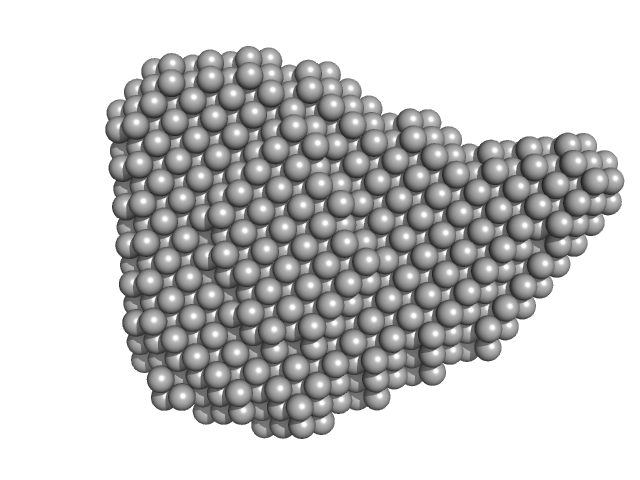
|
| Sample: |
Efa1/LifA protein (C1480A mutant) monomer, 367 kDa Escherichia coli O127:H6 … protein
|
| Buffer: |
50 mM Bis-Tris, 100 mM NaCl, 3 % (v/v) glycerol, pH: 5.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jun 29
|
Activity of lymphostatin, a lymphocyte inhibitory virulence factor of pathogenic Escherichia coli, is dependent on a cysteine protease motif.
J Mol Biol :167200 (2021)
Bease AG, Blackburn EA, Chintoan-Uta C, Webb S, Cassady-Cain RL, Stevens MP
|
| RgGuinier |
5.8 |
nm |
| Dmax |
19.3 |
nm |
| VolumePorod |
560 |
nm3 |
|
|
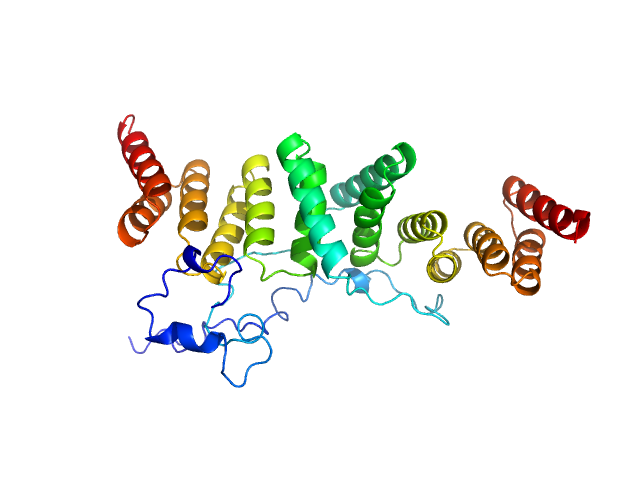
UniProt ID: P0A2U4 (2-155) Chaperone protein IpgC
|
|
|
|
| Sample: |
Chaperone protein IpgC dimer, 39 kDa Shigella flexneri protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2012 Dec 10
|
Structural Insights of Shigella Translocator IpaB and Its Chaperone IpgC in Solution
Frontiers in Cellular and Infection Microbiology 11 (2021)
Ferrari M, Charova S, Sansonetti P, Mylonas E, Gazi A
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
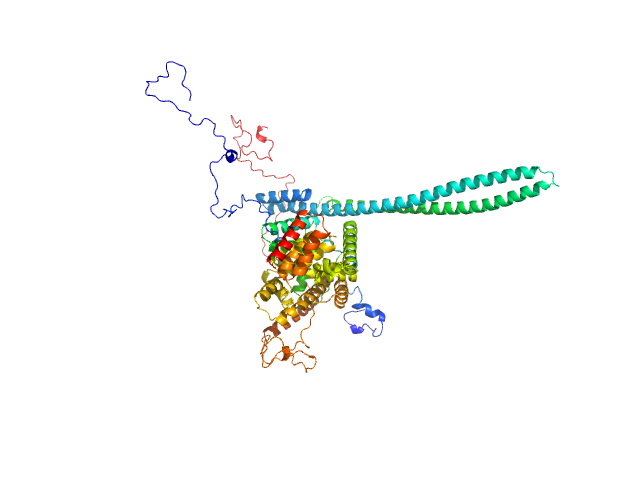
UniProt ID: P0A2U4 (2-155) T3SS Chaperone protein IpgC
UniProt ID: P18011 (1-580) Invasin IpaB
|
|
|
|
| Sample: |
T3SS Chaperone protein IpgC monomer, 20 kDa Shigella flexneri protein
Invasin IpaB monomer, 62 kDa Shigella flexneri protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2012 Dec 10
|
Structural Insights of Shigella Translocator IpaB and Its Chaperone IpgC in Solution
Frontiers in Cellular and Infection Microbiology 11 (2021)
Ferrari M, Charova S, Sansonetti P, Mylonas E, Gazi A
|
| RgGuinier |
3.9 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
165 |
nm3 |
|
|
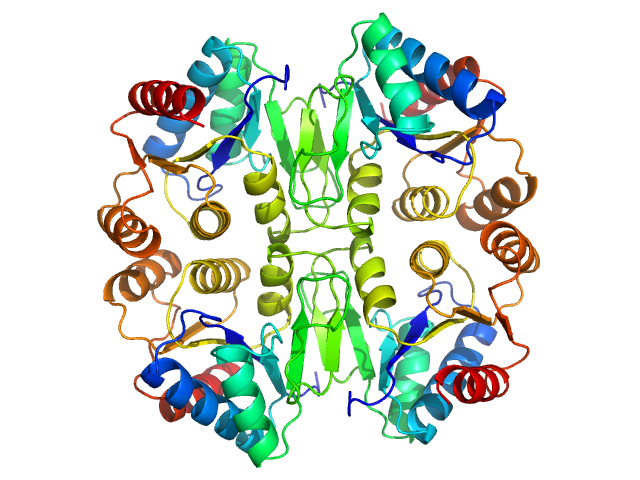
UniProt ID: B7K4Z0 (1-163) p450 cytochrome, putative (Moco carrier protein)
|
|
|
|
| Sample: |
P450 cytochrome, putative (Moco carrier protein) tetramer, 75 kDa Rippkaea orientalis (strain … protein
|
| Buffer: |
100 mM Tris-HCl, 300 mM NaCl, 2 %(v/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Feb 5
|
The structure of the Moco carrier protein from Rippkaea orientalis.
Acta Crystallogr F Struct Biol Commun 76(Pt 9):453-463 (2020)
Krausze J, Hercher TW, Archna A, Kruse T
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
107 |
nm3 |
|
|