UniProt ID: A0A1P6BP23 (1-213) dTMP kinase
|
|
|
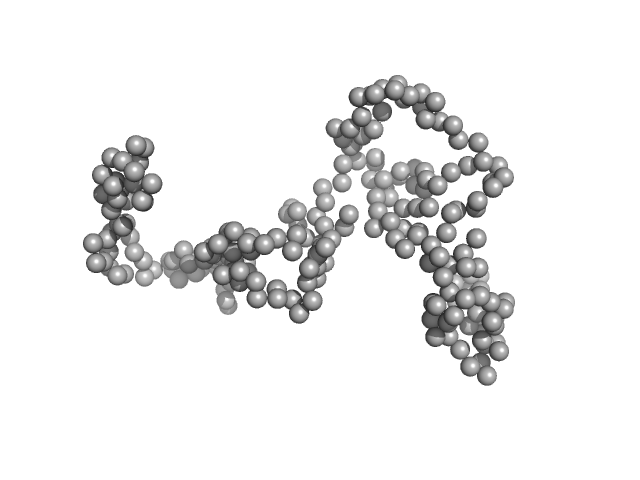
|
| Sample: |
DTMP kinase dimer, 57 kDa Brugia malayi protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2018 Oct 7
|
Crystal Structure of the Brugia malayi Thymidylate Kinase-dTMP Complex and Small Angle X-ray Scattering Experiments Identifies Changes in the Dimeric Association Compared to the Human Homolog
Crystallography Reports 68(7):1150-1158 (2024)
Vishwakarma J, Sharma V, Kumar S, Ramachandran R
|
| RgGuinier |
3.0 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
81 |
nm3 |
|
|
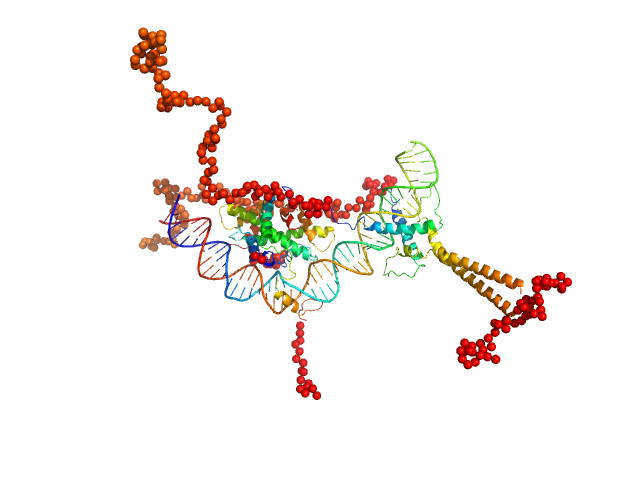
UniProt ID: P22415 (106-310) Upstream stimulatory factor 1
UniProt ID: P23511 (262-332) Nuclear transcription factor Y subunit alpha
UniProt ID: P25208 (52-143) Nuclear transcription factor Y subunit beta
UniProt ID: Q13952 (27-120) Nuclear transcription factor Y subunit gamma
UniProt ID: None (None-None) DNA 48bp
|
|
|
|
| Sample: |
Upstream stimulatory factor 1 dimer, 50 kDa Homo sapiens protein
Nuclear transcription factor Y subunit alpha monomer, 10 kDa Homo sapiens protein
Nuclear transcription factor Y subunit beta monomer, 11 kDa Homo sapiens protein
Nuclear transcription factor Y subunit gamma monomer, 11 kDa Homo sapiens protein
DNA 48bp monomer, 30 kDa DNA
|
| Buffer: |
100 mM cacodylate buffer, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 14
|
The USR domain of USF1 mediates NF-Y interactions and cooperative DNA binding
International Journal of Biological Macromolecules (2021)
Bernardini A, Lorenzo M, Chaves-Sanjuan A, Swuec P, Pigni M, Saad D, Konarev P, Graewert M, Valentini E, Svergun D, Nardini M, Mantovani R, Gnesutta N
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
155 |
nm3 |
|
|
UniProt ID: P22415 (106-310) Upstream stimulatory factor 1
UniProt ID: P23511 (262-332) Nuclear transcription factor Y subunit alpha
UniProt ID: P25208 (52-143) Nuclear transcription factor Y subunit beta
UniProt ID: Q13952 (27-120) Nuclear transcription factor Y subunit gamma
UniProt ID: None (None-None) DNA 50bp
|
|
|

|
| Sample: |
Upstream stimulatory factor 1 dimer, 50 kDa Homo sapiens protein
Nuclear transcription factor Y subunit alpha monomer, 10 kDa Homo sapiens protein
Nuclear transcription factor Y subunit beta monomer, 11 kDa Homo sapiens protein
Nuclear transcription factor Y subunit gamma monomer, 11 kDa Homo sapiens protein
DNA 50bp monomer, 31 kDa DNA
|
| Buffer: |
100 mM cacodylate buffer, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 14
|
The USR domain of USF1 mediates NF-Y interactions and cooperative DNA binding
International Journal of Biological Macromolecules (2021)
Bernardini A, Lorenzo M, Chaves-Sanjuan A, Swuec P, Pigni M, Saad D, Konarev P, Graewert M, Valentini E, Svergun D, Nardini M, Mantovani R, Gnesutta N
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
165 |
nm3 |
|
|
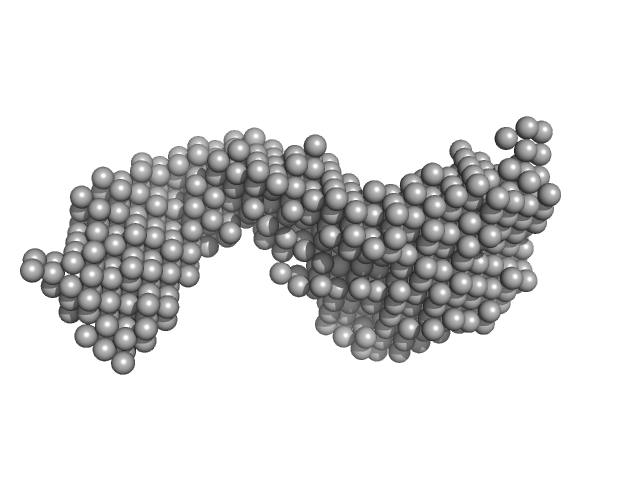
UniProt ID: P61244 (1-160) Protein max (Isoform 2, short, 13-21: missing)
UniProt ID: P23511 (262-332) Nuclear transcription factor Y subunit alpha
UniProt ID: P25208 (52-143) Nuclear transcription factor Y subunit beta
UniProt ID: Q13952 (27-120) Nuclear transcription factor Y subunit gamma
UniProt ID: None (None-None) DNA 48bp
|
|
|
|
| Sample: |
Protein max (Isoform 2, short, 13-21: missing) dimer, 39 kDa Homo sapiens protein
Nuclear transcription factor Y subunit alpha monomer, 10 kDa Homo sapiens protein
Nuclear transcription factor Y subunit beta monomer, 11 kDa Homo sapiens protein
Nuclear transcription factor Y subunit gamma monomer, 11 kDa Homo sapiens protein
DNA 48bp monomer, 30 kDa DNA
|
| Buffer: |
100 mM cacodylate buffer, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2012 Dec 10
|
The USR domain of USF1 mediates NF-Y interactions and cooperative DNA binding
International Journal of Biological Macromolecules (2021)
Bernardini A, Lorenzo M, Chaves-Sanjuan A, Swuec P, Pigni M, Saad D, Konarev P, Graewert M, Valentini E, Svergun D, Nardini M, Mantovani R, Gnesutta N
|
| RgGuinier |
5.2 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
151 |
nm3 |
|
|
UniProt ID: Q7MJG0 (None-None) Type I restriction-modification system methyltransferase subunit
UniProt ID: P03775 (None-None) Protein Ocr
|
|
|

|
| Sample: |
Type I restriction-modification system methyltransferase subunit dimer, 144 kDa Vibrio vulnificus (strain … protein
Protein Ocr dimer, 28 kDa Escherichia phage T7 protein
|
| Buffer: |
20 mM Tris-HCl,150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 4C, Pohang Accelerator Laboratory on 2020 Apr 22
|
Structural features of a minimal intact methyltransferase of a type I restriction-modification system.
Int J Biol Macromol (2022)
Seo PW, Hofmann A, Kim JH, Hwangbo SA, Kim JH, Kim JW, Huynh TYL, Choy HE, Kim SJ, Lee J, Lee JO, Jin KS, Park SY, Kim JS
|
| RgGuinier |
4.2 |
nm |
| Dmax |
15.9 |
nm |
| VolumePorod |
354 |
nm3 |
|
|
UniProt ID: P11142 (1-646) Heat shock cognate 71 kDa protein
|
|
|
|
| Sample: |
Heat shock cognate 71 kDa protein monomer, 71 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris HCl, 50 mM NaCl, 5 mM Sodium Phosphate, 5 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS1 Beamline, Brazilian Synchrotron Light Laboratory on 2018 May 18
|
Structural, thermodynamic and functional studies of human 71 kDa heat shock cognate protein (HSPA8/hHsc70)
Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics :140719 (2021)
Silva N, de Camargo Rodrigues L, Dores-Silva P, Montanari C, Ramos C, Barbosa L, Borges J
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
111 |
nm3 |
|
|
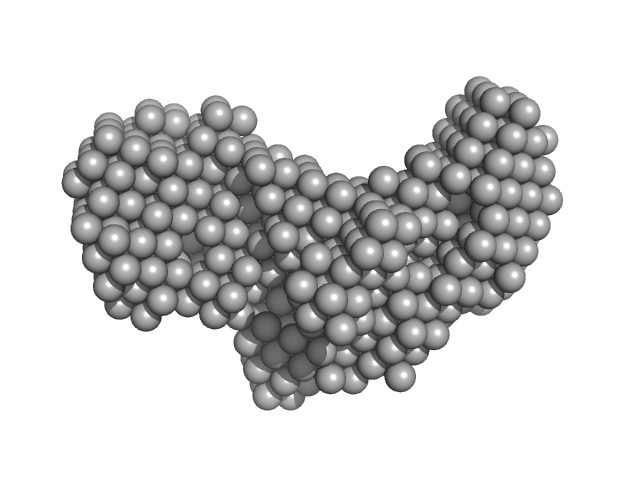
UniProt ID: P0DMV8 (1-586) Heat shock 70 kDa protein 1A
|
|
|
|
| Sample: |
Heat shock 70 kDa protein 1A monomer, 70 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris HCl, 50 mM NaCl, 5 mM Sodium Phosphate, 5 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS1 Beamline, Brazilian Synchrotron Light Laboratory on 2018 May 18
|
Structural, thermodynamic and functional studies of human 71 kDa heat shock cognate protein (HSPA8/hHsc70)
Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics :140719 (2021)
Silva N, de Camargo Rodrigues L, Dores-Silva P, Montanari C, Ramos C, Barbosa L, Borges J
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
138 |
nm3 |
|
|
UniProt ID: P0DKX7 (None-None) hybrid RTX-1 construct (amino acids 1132-1294 and 1562-1681 of CyaA)
|
|
|

|
| Sample: |
Hybrid RTX-1 construct (amino acids 1132-1294 and 1562-1681 of CyaA) monomer, 30 kDa Bordetella pertussis protein
|
| Buffer: |
10 mM Tris HCl, 150 mM NaCl, 10 mM CaCl₂, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Dec 1
|
Almost half of the RTX domain is dispensable for complement receptor 3 binding and cell-invasive activity of the adenylate cyclase toxin.
J Biol Chem :100833 (2021)
Espinosa-Vinals CA, Masin J, Holubova J, Stanek O, Jurnecka D, Osicka R, Sebo P, Bumba L
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
UniProt ID: P0DKX7 (None-None) hybrid RTX-2 construct (amino acids 1132-1303 and 1562-1681 of CyaA)
|
|
|

|
| Sample: |
Hybrid RTX-2 construct (amino acids 1132-1303 and 1562-1681 of CyaA) monomer, 31 kDa Bordetella pertussis protein
|
| Buffer: |
10 mM Tris HCl, 150 mM NaCl, 10 mM CaCl₂, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Dec 1
|
Almost half of the RTX domain is dispensable for complement receptor 3 binding and cell-invasive activity of the adenylate cyclase toxin.
J Biol Chem :100833 (2021)
Espinosa-Vinals CA, Masin J, Holubova J, Stanek O, Jurnecka D, Osicka R, Sebo P, Bumba L
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
37 |
nm3 |
|
|
UniProt ID: P22303-1 (32-578) acetylcholinesterase
UniProt ID: P22303-1 (32-578) acetylcholinesterase
|
|
|
|
| Sample: |
Acetylcholinesterase dimer, 120 kDa Homo sapiens protein
Acetylcholinesterase monomer, 60 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris/HCl, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2015 May 29
|
Covalent inhibition of hAChE by organophosphates causes homodimer dissociation through long-range allosteric effects.
J Biol Chem :101007 (2021)
Blumenthal DK, Cheng X, Fajer M, Ho KY, Rohrer J, Gerlits O, Taylor P, Juneja P, Kovalevsky A, Radić Z
|
| RgGuinier |
3.9 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
162 |
nm3 |
|
|