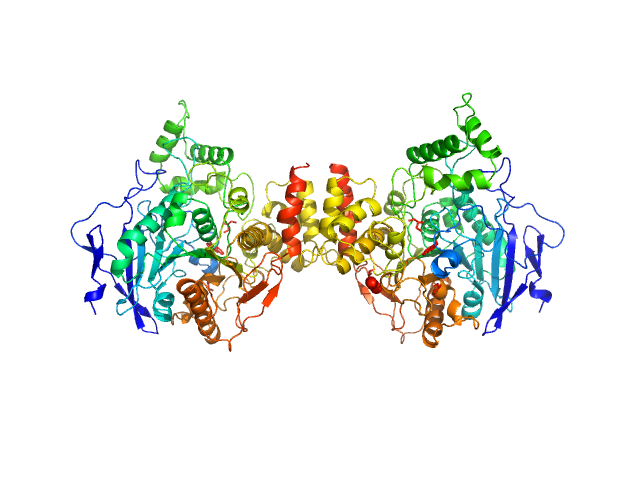
UniProt ID: P22303-1 (32-578) acetylcholinesterase
UniProt ID: P22303-1 (32-578) acetylcholinesterase
|
|
|
|
| Sample: |
Acetylcholinesterase dimer, 120 kDa Homo sapiens protein
Acetylcholinesterase monomer, 60 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris/HCl, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2015 May 29
|
Covalent inhibition of hAChE by organophosphates causes homodimer dissociation through long-range allosteric effects.
J Biol Chem :101007 (2021)
Blumenthal DK, Cheng X, Fajer M, Ho KY, Rohrer J, Gerlits O, Taylor P, Juneja P, Kovalevsky A, Radić Z
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
142 |
nm3 |
|
|
UniProt ID: Q15796 (1-463) Mothers against decapentaplegic homolog 2 (C-terminus phosphomimetic mutant)
|
|
|
|
| Sample: |
Mothers against decapentaplegic homolog 2 (C-terminus phosphomimetic mutant), 53 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Apr 4
|
Conformational landscape of multidomain SMAD proteins
Computational and Structural Biotechnology Journal (2021)
Gomes T, Martin-Malpartida P, Ruiz L, Aragón E, Cordeiro T, Macias M
|
| RgGuinier |
5.1 |
nm |
| Dmax |
20.0 |
nm |
|
|
UniProt ID: Q15796 (1-463) Mothers against decapentaplegic homolog 2 (C-terminus phosphomimetic mutant)
|
|
|
|
| Sample: |
Mothers against decapentaplegic homolog 2 (C-terminus phosphomimetic mutant), 53 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Apr 4
|
Conformational landscape of multidomain SMAD proteins
Computational and Structural Biotechnology Journal (2021)
Gomes T, Martin-Malpartida P, Ruiz L, Aragón E, Cordeiro T, Macias M
|
| RgGuinier |
5.2 |
nm |
| Dmax |
20.0 |
nm |
|
|
UniProt ID: Q15796 (1-463) Mothers against decapentaplegic homolog 2 (C-terminus phosphomimetic mutant)
|
|
|
|
| Sample: |
Mothers against decapentaplegic homolog 2 (C-terminus phosphomimetic mutant), 53 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Apr 4
|
Conformational landscape of multidomain SMAD proteins
Computational and Structural Biotechnology Journal (2021)
Gomes T, Martin-Malpartida P, Ruiz L, Aragón E, Cordeiro T, Macias M
|
| RgGuinier |
5.3 |
nm |
| Dmax |
20.0 |
nm |
|
|
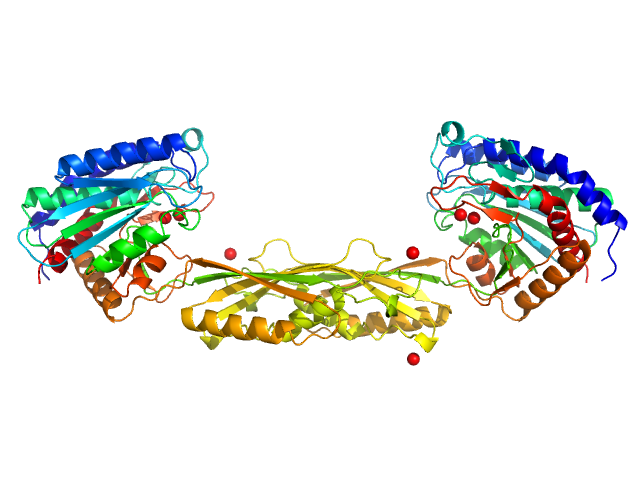
UniProt ID: P06621 (89-415) Carboxypeptidase G2 (circular permutant CP-N89) K177A
|
|
|
|
| Sample: |
Carboxypeptidase G2 (circular permutant CP-N89) K177A dimer, 85 kDa Pseudomonas sp. (strain … protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Jun 6
|
Massively parallel, computationally guided design of a proenzyme.
Proc Natl Acad Sci U S A 119(15):e2116097119 (2022)
Yachnin BJ, Azouz LR, White RE 3rd, Minetti CASA, Remeta DP, Tan VM, Drake JM, Khare SD
|
| RgGuinier |
3.6 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
105 |
nm3 |
|
|
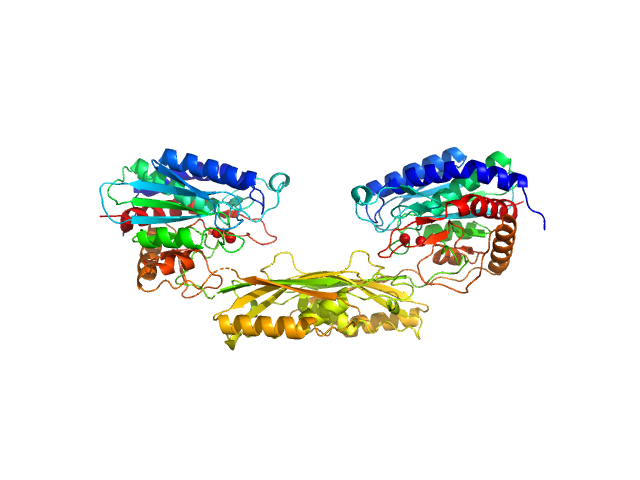
UniProt ID: P06621 (89-415) Carboxypeptidase G2 (circular permutant CP-N89) K177A
UniProt ID: None (None-None) Methotrexate
|
|
|
|
| Sample: |
Carboxypeptidase G2 (circular permutant CP-N89) K177A dimer, 85 kDa Pseudomonas sp. (strain … protein
Methotrexate dimer, 1 kDa
|
| Buffer: |
50 mM Tris, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Jun 6
|
Massively parallel, computationally guided design of a proenzyme.
Proc Natl Acad Sci U S A 119(15):e2116097119 (2022)
Yachnin BJ, Azouz LR, White RE 3rd, Minetti CASA, Remeta DP, Tan VM, Drake JM, Khare SD
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
104 |
nm3 |
|
|
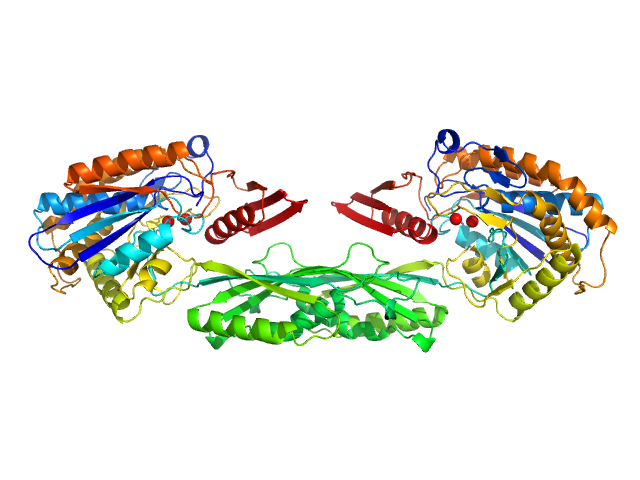
UniProt ID: P06621 (89-415) Pro-Carboxypeptidase G2 (circular permutant CP-N89) K177A Design 1
|
|
|
|
| Sample: |
Pro-Carboxypeptidase G2 (circular permutant CP-N89) K177A Design 1 dimer, 100 kDa Pseudomonas sp. (strain … protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 May 8
|
Massively parallel, computationally guided design of a proenzyme.
Proc Natl Acad Sci U S A 119(15):e2116097119 (2022)
Yachnin BJ, Azouz LR, White RE 3rd, Minetti CASA, Remeta DP, Tan VM, Drake JM, Khare SD
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.4 |
nm |
| VolumePorod |
125 |
nm3 |
|
|
UniProt ID: None (None-None) Methotrexate
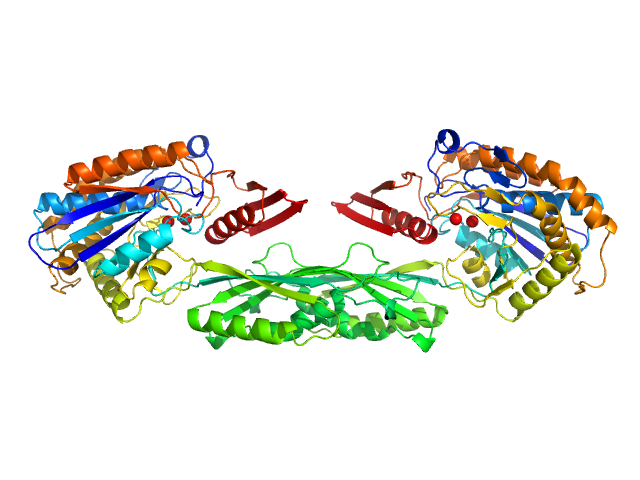
UniProt ID: P06621 (89-415) Pro-Carboxypeptidase G2 (circular permutant CP-N89) K177A Design 1
|
|
|
|
| Sample: |
Methotrexate dimer, 1 kDa
Pro-Carboxypeptidase G2 (circular permutant CP-N89) K177A Design 1 dimer, 100 kDa Pseudomonas sp. (strain … protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 May 8
|
Massively parallel, computationally guided design of a proenzyme.
Proc Natl Acad Sci U S A 119(15):e2116097119 (2022)
Yachnin BJ, Azouz LR, White RE 3rd, Minetti CASA, Remeta DP, Tan VM, Drake JM, Khare SD
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
123 |
nm3 |
|
|
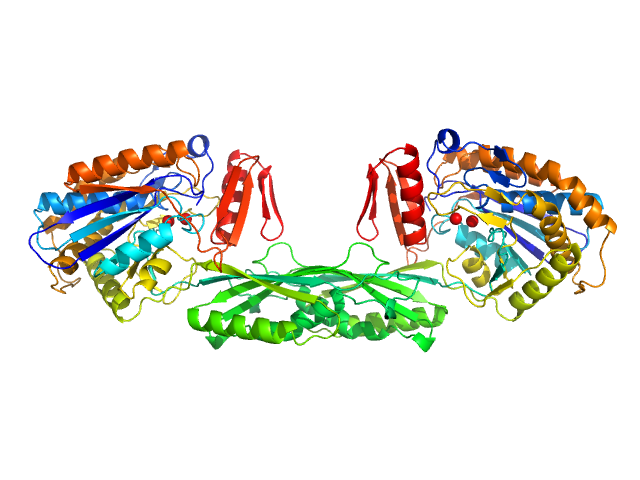
UniProt ID: P06621 (89-415) Pro-Carboxypeptidase G2 (circular permutant CP-N89) K177A Design 2
|
|
|
|
| Sample: |
Pro-Carboxypeptidase G2 (circular permutant CP-N89) K177A Design 2 dimer, 100 kDa Pseudomonas sp. (strain … protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 May 8
|
Massively parallel, computationally guided design of a proenzyme.
Proc Natl Acad Sci U S A 119(15):e2116097119 (2022)
Yachnin BJ, Azouz LR, White RE 3rd, Minetti CASA, Remeta DP, Tan VM, Drake JM, Khare SD
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
126 |
nm3 |
|
|
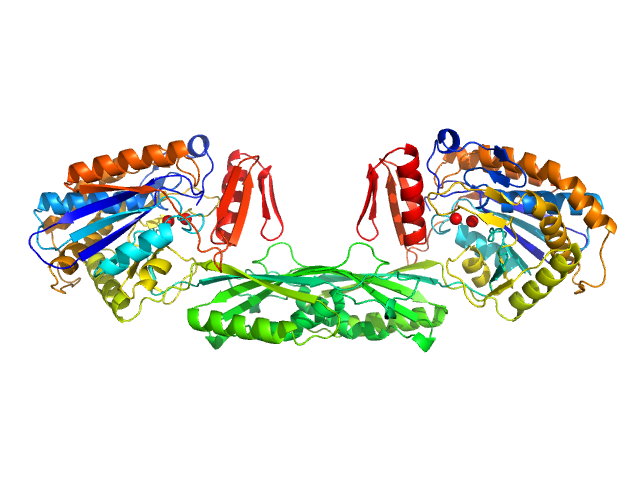
UniProt ID: None (None-None) Methotrexate
UniProt ID: P06621 (89-415) Pro-Carboxypeptidase G2 (circular permutant CP-N89) K177A Design 2
|
|
|
|
| Sample: |
Methotrexate dimer, 1 kDa
Pro-Carboxypeptidase G2 (circular permutant CP-N89) K177A Design 2 dimer, 100 kDa Pseudomonas sp. (strain … protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 May 8
|
Massively parallel, computationally guided design of a proenzyme.
Proc Natl Acad Sci U S A 119(15):e2116097119 (2022)
Yachnin BJ, Azouz LR, White RE 3rd, Minetti CASA, Remeta DP, Tan VM, Drake JM, Khare SD
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
125 |
nm3 |
|
|