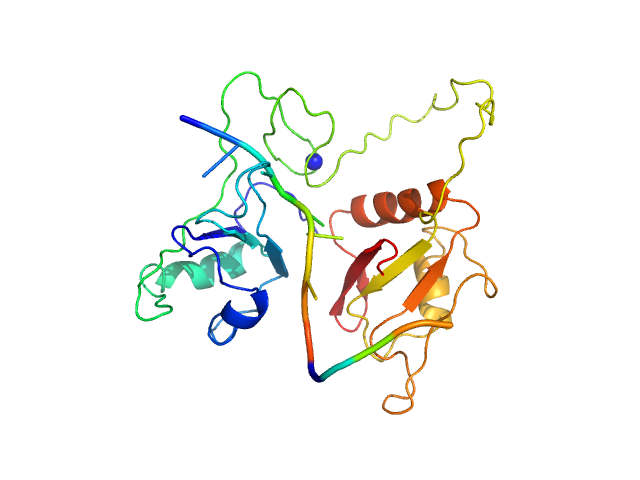
UniProt ID: P52756 (94-315) RNA Binding Motif protein 5 (I107T, C191G)
UniProt ID: None (None-None) Caspase-2 derived RNA GGCU_12
|
|
|
|
| Sample: |
RNA Binding Motif protein 5 (I107T, C191G) monomer, 26 kDa Homo sapiens protein
Caspase-2 derived RNA GGCU_12 monomer, 4 kDa RNA
|
| Buffer: |
20 mM MES, 100 mM NaCl, 1 mM DTT, pH: 6.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, SFB 1035, Technische Universität München on 2017 Mar 1
|
Structural basis for specific RNA recognition by the alternative splicing factor RBM5.
Nat Commun 14(1):4233 (2023)
Soni K, Jagtap PKA, Martínez-Lumbreras S, Bonnal S, Geerlof A, Stehle R, Simon B, Valcárcel J, Sattler M
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
42 |
nm3 |
|
|
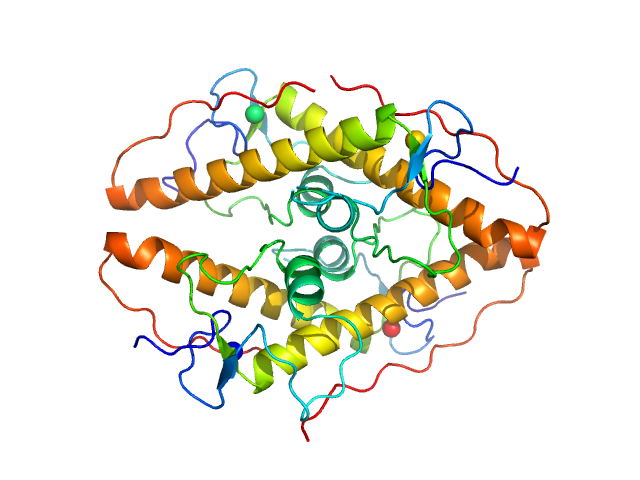
UniProt ID: Q95RQ8 (1-111) LD15650p (Pita, isoform A)
|
|
|
|
| Sample: |
LD15650p (Pita, isoform A) tetramer, 52 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 5 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jul 8
|
Structural insights into highly similar spatial organization of zinc-finger associated domains with a very low sequence similarity.
Structure (2022)
Bonchuk AN, Boyko KM, Nikolaeva AY, Burtseva AD, Popov VO, Georgiev PG
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
62 |
nm3 |
|
|
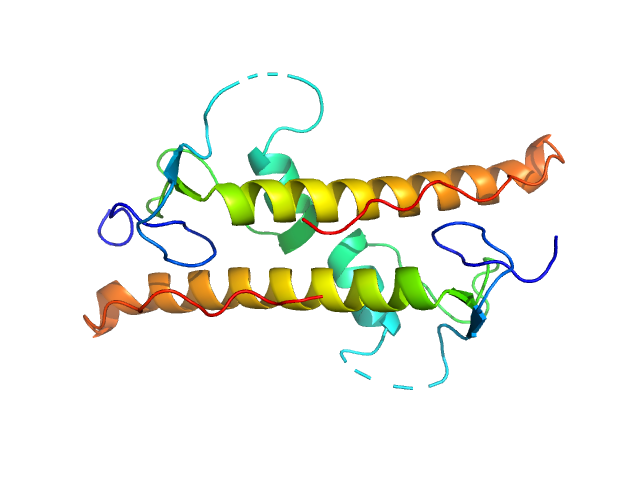
UniProt ID: Q95RQ8 (1-111) LD15650p (Pita, isoform A; L45A)
|
|
|
|
| Sample: |
LD15650p (Pita, isoform A; L45A) dimer, 26 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 5 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Dec 17
|
Structural insights into highly similar spatial organization of zinc-finger associated domains with a very low sequence similarity.
Structure (2022)
Bonchuk AN, Boyko KM, Nikolaeva AY, Burtseva AD, Popov VO, Georgiev PG
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
35 |
nm3 |
|
|
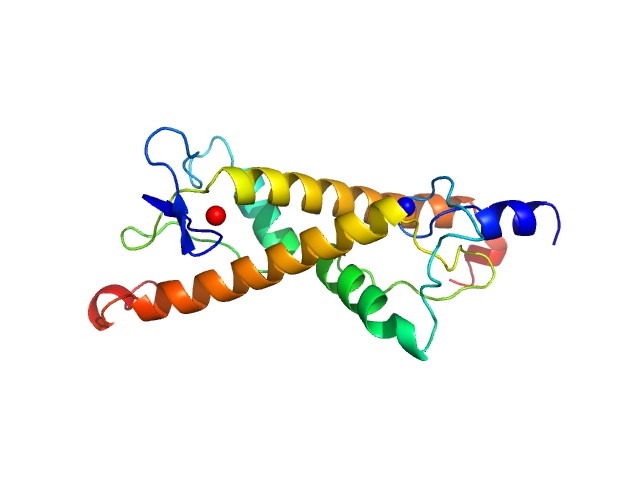
UniProt ID: Q9VHM3 (1-98) LD30467p (Motif 1 binding protein)
|
|
|
|
| Sample: |
LD30467p (Motif 1 binding protein) dimer, 22 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Bis-Tris-Propane, 100 mM NaCl, 5 mM DTT, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Dec 17
|
Structural insights into highly similar spatial organization of zinc-finger associated domains with a very low sequence similarity.
Structure (2022)
Bonchuk AN, Boyko KM, Nikolaeva AY, Burtseva AD, Popov VO, Georgiev PG
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
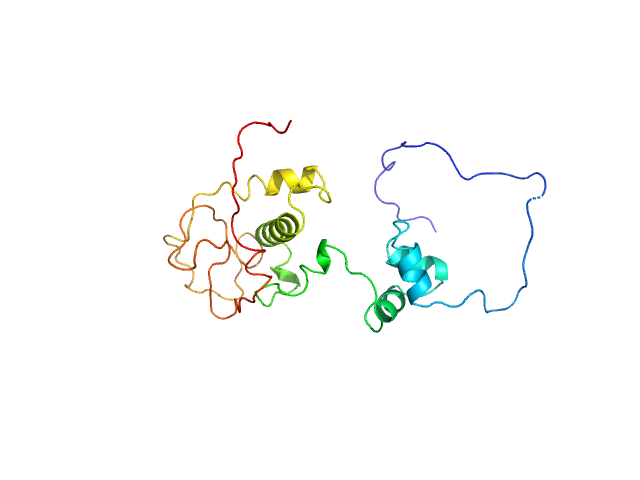
UniProt ID: A0A2D1GKF7 (1-183) Immunity repressor
|
|
|
|
| Sample: |
Immunity repressor monomer, 24 kDa Mycobacterium phage TipsytheTRex protein
|
| Buffer: |
20 mM Tris pH 7.5, 0.5 M NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Sep 10
|
A monomeric mycobacteriophage immunity repressor utilizes two domains to recognize an asymmetric DNA sequence.
Nat Commun 13(1):4105 (2022)
McGinnis RJ, Brambley CA, Stamey B, Green WC, Gragg KN, Cafferty ER, Terwilliger TC, Hammel M, Hollis TJ, Miller JM, Gainey MD, Wallen JR
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
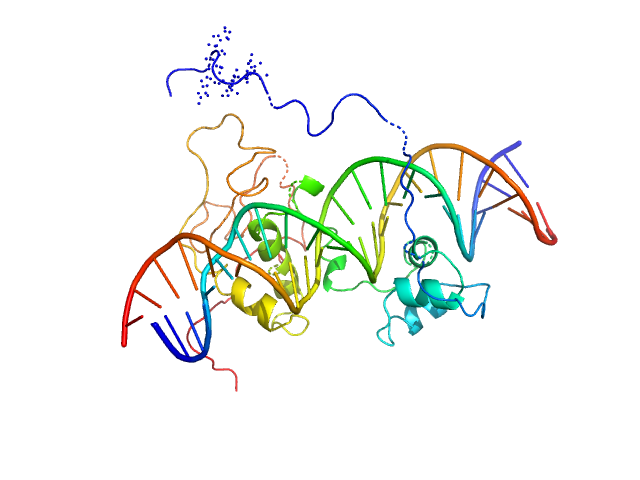
UniProt ID: A0A2D1GKF7 (1-183) Immunity repressor
UniProt ID: None (None-None) 24mer dsDNA
|
|
|
|
| Sample: |
Immunity repressor monomer, 21 kDa Mycobacterium phage TipsytheTRex protein
24mer dsDNA monomer, 15 kDa DNA
|
| Buffer: |
20 mM Tris pH 7.5, 0.5 M NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Sep 10
|
A monomeric mycobacteriophage immunity repressor utilizes two domains to recognize an asymmetric DNA sequence.
Nat Commun 13(1):4105 (2022)
McGinnis RJ, Brambley CA, Stamey B, Green WC, Gragg KN, Cafferty ER, Terwilliger TC, Hammel M, Hollis TJ, Miller JM, Gainey MD, Wallen JR
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
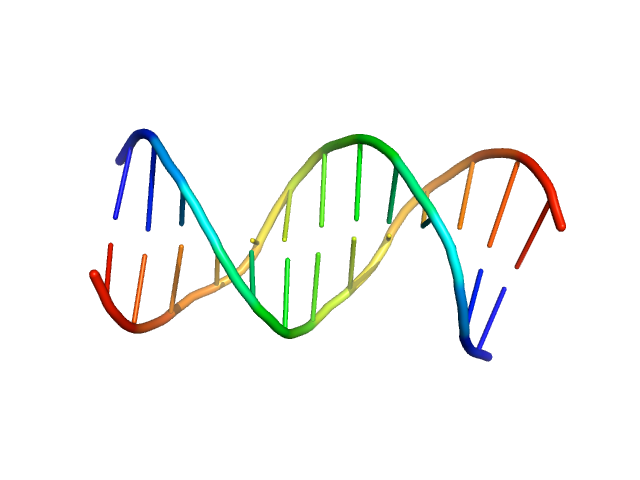
UniProt ID: A0A2D1GKF7 (1-183) Immunity repressor
UniProt ID: None (None-None) 13mer dsDNA
|
|
|
|
| Sample: |
Immunity repressor monomer, 21 kDa Mycobacterium phage TipsytheTRex protein
13mer dsDNA monomer, 8 kDa DNA
|
| Buffer: |
20 mM Tris pH 7.5, 0.5 M NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Dec 10
|
A monomeric mycobacteriophage immunity repressor utilizes two domains to recognize an asymmetric DNA sequence.
Nat Commun 13(1):4105 (2022)
McGinnis RJ, Brambley CA, Stamey B, Green WC, Gragg KN, Cafferty ER, Terwilliger TC, Hammel M, Hollis TJ, Miller JM, Gainey MD, Wallen JR
|
| RgGuinier |
2.0 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
UniProt ID: Q6UWE0 (1-723) E3 ubiquitin-protein ligase LRSAM1
|
|
|

|
| Sample: |
E3 ubiquitin-protein ligase LRSAM1 monomer, 84 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2019 Dec 7
|
Leucine Rich Repeat And Sterile Alpha Motif Containing 1 (LRSAM1)
Si Hoon Park
|
| RgGuinier |
6.1 |
nm |
| Dmax |
19.4 |
nm |
| VolumePorod |
250 |
nm3 |
|
|
UniProt ID: Q6UWE0 (1-632) E3 ubiquitin-protein ligase LRSAM1 - ΔPTAP/PSAP-RING
|
|
|

|
| Sample: |
E3 ubiquitin-protein ligase LRSAM1 - ΔPTAP/PSAP-RING monomer, 74 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl pH 8.0, 300 mM NaCl, 1 mM TCEP, 5% w/v glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 4C, Pohang Accelerator Laboratory on 2017 Oct 24
|
Leucine Rich Repeat And Sterile Alpha Motif Containing 1 (LRSAM1)
Si Hoon Park
|
| RgGuinier |
5.5 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
322 |
nm3 |
|
|
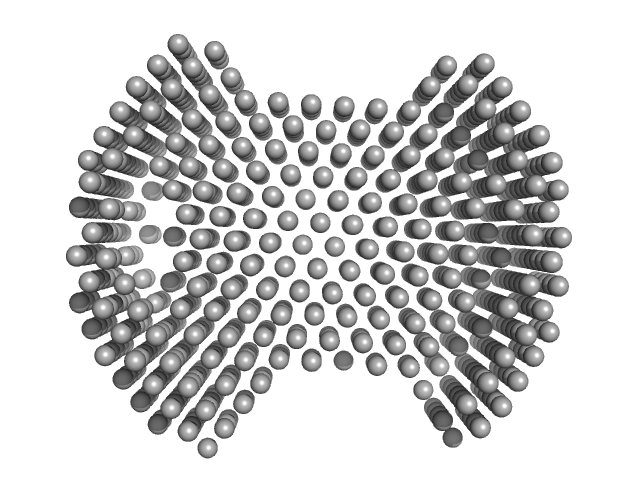
UniProt ID: Q6UWE0 (1-307) E3 ubiquitin-protein ligase LRSAM1 - LRR containing N-terminal domain
|
|
|
|
| Sample: |
E3 ubiquitin-protein ligase LRSAM1 - LRR containing N-terminal domain monomer, 35 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl pH 8.0, 300 mM NaCl, 1 mM TCEP, 5% w/v glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 4C, Pohang Accelerator Laboratory on 2017 Oct 23
|
Leucine Rich Repeat And Sterile Alpha Motif Containing 1 (LRSAM1)
Si Hoon Park
|
| RgGuinier |
3.4 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
105 |
nm3 |
|
|