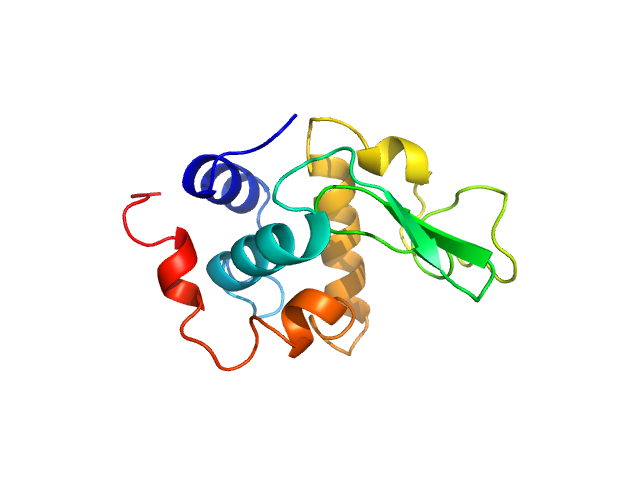
UniProt ID: P00698 (19-147) Lysozyme C
|
|
|
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
40 mM NaOAc pH 3.8, 150 mM NaCl, pH: 3.8 |
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2014 May 2
|
Visualizing how inclusion of higher reciprocal space in SWAXS data analysis improves shape restoration of biomolecules: case of lysozyme.
J Biomol Struct Dyn :1-15 (2021)
Ashish
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.6 |
nm |
|
|
UniProt ID: P00698 (19-147) Lysozyme C
|
|
|

|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
40 mM NaOAc pH 3.8, 150 mM NaCl, pH: 3.8 |
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2014 May 2
|
Visualizing how inclusion of higher reciprocal space in SWAXS data analysis improves shape restoration of biomolecules: case of lysozyme.
J Biomol Struct Dyn :1-15 (2021)
Ashish
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.2 |
nm |
|
|
UniProt ID: P00698 (19-147) Lysozyme C
|
|
|

|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
40 mM NaOAc pH 3.8, 150 mM NaCl, pH: 3.8 |
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2014 May 2
|
Visualizing how inclusion of higher reciprocal space in SWAXS data analysis improves shape restoration of biomolecules: case of lysozyme.
J Biomol Struct Dyn :1-15 (2021)
Ashish
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.2 |
nm |
|
|
UniProt ID: P00698 (19-147) Lysozyme C
|
|
|

|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
40 mM NaOAc pH 3.8, 150 mM NaCl, pH: 3.8 |
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2014 May 2
|
Visualizing how inclusion of higher reciprocal space in SWAXS data analysis improves shape restoration of biomolecules: case of lysozyme.
J Biomol Struct Dyn :1-15 (2021)
Ashish
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.2 |
nm |
|
|
UniProt ID: P00698 (19-147) Lysozyme C
|
|
|
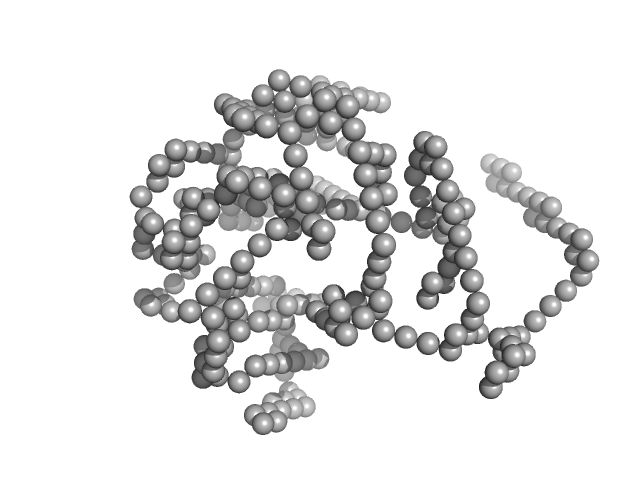
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
40 mM NaOAc pH 3.8, 150 mM NaCl, pH: 3.8 |
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2014 May 2
|
Visualizing how inclusion of higher reciprocal space in SWAXS data analysis improves shape restoration of biomolecules: case of lysozyme.
J Biomol Struct Dyn :1-15 (2021)
Ashish
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.2 |
nm |
|
|
UniProt ID: Q7TSU7 (21-506) Kin of IRRE-like protein 2
|
|
|
|
| Sample: |
Kin of IRRE-like protein 2 dimer, 106 kDa Mus musculus protein
|
| Buffer: |
10 mM HEPES pH 7.2, 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Aug 3
|
Molecular and structural basis of olfactory sensory neuron axon coalescence by Kirrel receptors.
Cell Rep 37(5):109940 (2021)
Wang J, Vaddadi N, Pak JS, Park Y, Quilez S, Roman CA, Dumontier E, Thornton JW, Cloutier JF, Özkan E
|
| RgGuinier |
8.9 |
nm |
| Dmax |
39.0 |
nm |
|
|
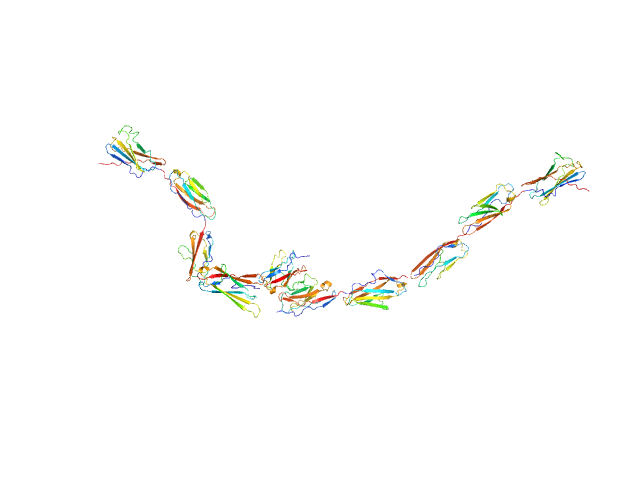
UniProt ID: Q8BR86 (47-517) Kin of IRRE-like protein 3
|
|
|
|
| Sample: |
Kin of IRRE-like protein 3 dimer, 106 kDa Mus musculus protein
|
| Buffer: |
10 mM HEPES pH 7.2, 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Aug 3
|
Molecular and structural basis of olfactory sensory neuron axon coalescence by Kirrel receptors.
Cell Rep 37(5):109940 (2021)
Wang J, Vaddadi N, Pak JS, Park Y, Quilez S, Roman CA, Dumontier E, Thornton JW, Cloutier JF, Özkan E
|
| RgGuinier |
9.3 |
nm |
| Dmax |
34.5 |
nm |
|
|
UniProt ID: Q8BR86 (47-517) Kin of IRRE-like protein 3 (Q128A)
|
|
|
|
| Sample: |
Kin of IRRE-like protein 3 (Q128A) monomer, 53 kDa Mus musculus protein
|
| Buffer: |
10 mM HEPES pH 7.2, 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Aug 3
|
Molecular and structural basis of olfactory sensory neuron axon coalescence by Kirrel receptors.
Cell Rep 37(5):109940 (2021)
Wang J, Vaddadi N, Pak JS, Park Y, Quilez S, Roman CA, Dumontier E, Thornton JW, Cloutier JF, Özkan E
|
| RgGuinier |
5.4 |
nm |
| Dmax |
21.3 |
nm |
|
|
UniProt ID: P52756 (94-210) RNA-binding protein 5 (I107T, C191G)
|
|
|
|
| Sample: |
RNA-binding protein 5 (I107T, C191G) monomer, 14 kDa Homo sapiens protein
|
| Buffer: |
20 mM MES, 400 mM NaCl, 1 mM DTT, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Sep 26
|
Structural basis for specific RNA recognition by the alternative splicing factor RBM5.
Nat Commun 14(1):4233 (2023)
Soni K, Jagtap PKA, Martínez-Lumbreras S, Bonnal S, Geerlof A, Stehle R, Simon B, Valcárcel J, Sattler M
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.6 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
UniProt ID: P52756 (94-315) RNA Binding Motif protein 5 (I107T, C191G)
|
|
|
|
| Sample: |
RNA Binding Motif protein 5 (I107T, C191G) monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
20 mM MES, 400 mM NaCl, 1 mM DTT, pH: 6.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, SFB 1035, Technische Universität München on 2017 Feb 9
|
Structural basis for specific RNA recognition by the alternative splicing factor RBM5.
Nat Commun 14(1):4233 (2023)
Soni K, Jagtap PKA, Martínez-Lumbreras S, Bonnal S, Geerlof A, Stehle R, Simon B, Valcárcel J, Sattler M
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
36 |
nm3 |
|
|