UniProt ID: Q6UWE0 (561-632) E3 ubiquitin-protein ligase LRSAM1 - SAM domain
|
|
|

|
| Sample: |
E3 ubiquitin-protein ligase LRSAM1 - SAM domain monomer, 8 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at 4C, Pohang Accelerator Laboratory on 2019 Apr 4
|
Leucine Rich Repeat And Sterile Alpha Motif Containing 1 (LRSAM1)
Si Hoon Park
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.3 |
nm |
| VolumePorod |
11 |
nm3 |
|
|
UniProt ID: Q6UWE0 (80-171) E3 ubiquitin-protein ligase LRSAM1
|
|
|

|
| Sample: |
E3 ubiquitin-protein ligase LRSAM1 monomer, 30 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at 4C, Pohang Accelerator Laboratory on 2019 Apr 4
|
Leucine Rich Repeat And Sterile Alpha Motif Containing 1 (LRSAM1)
Si Hoon Park
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
UniProt ID: P00698 (None-None) lysozyme amyloid fibril
|
|
|
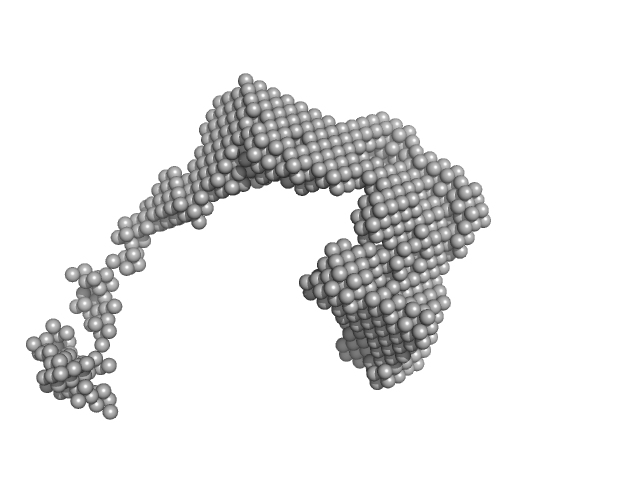
|
| Sample: |
Lysozyme amyloid fibril, 1 kDa Gallus gallus protein
|
| Buffer: |
0.2 M glycine-HCl, 80 mM NaCl, pH: 2.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 29
|
Dependence of the Nanoscale Composite Morphology of Fe3O4 Nanoparticle-Infused Lysozyme Amyloid Fibrils on Timing of Infusion: A Combined SAXS and AFM Study
Molecules 26(16):4864 (2021)
Schroer M, Hu P, Tomasovicova N, Batkova M, Zakutanska K, Wu P, Kopcansky P
|
| RgGuinier |
38.4 |
nm |
| Dmax |
80.0 |
nm |
|
|
UniProt ID: P00698 (None-None) lysozyme amyloid fibril
UniProt ID: None (None-None) Fe3O4 nanoparticles; nominal diameter 10 nm (hydrodynamic diameter)
|
|
|
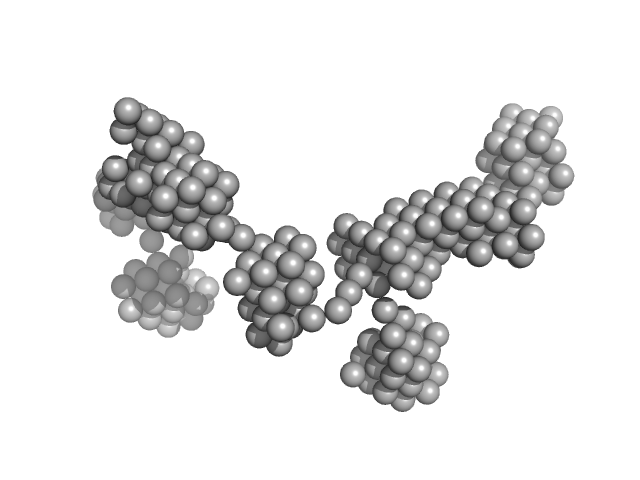
|
| Sample: |
Lysozyme amyloid fibril, 1 kDa Gallus gallus protein
Fe3O4 nanoparticles; nominal diameter 10 nm (hydrodynamic diameter) monomer, 1 kDa
|
| Buffer: |
0.2 M glycine-HCl, 80 mM NaCl, pH: 2.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 29
|
Dependence of the Nanoscale Composite Morphology of Fe3O4 Nanoparticle-Infused Lysozyme Amyloid Fibrils on Timing of Infusion: A Combined SAXS and AFM Study
Molecules 26(16):4864 (2021)
Schroer M, Hu P, Tomasovicova N, Batkova M, Zakutanska K, Wu P, Kopcansky P
|
| RgGuinier |
22.3 |
nm |
| Dmax |
70.0 |
nm |
|
|
UniProt ID: P00698 (None-None) lysozyme amyloid fibril
UniProt ID: None (None-None) Fe3O4 nanoparticles; nominal diameter 30 nm (hydrodynamic diameter)
|
|
|

|
| Sample: |
Lysozyme amyloid fibril, 1 kDa Gallus gallus protein
Fe3O4 nanoparticles; nominal diameter 30 nm (hydrodynamic diameter) monomer, 1 kDa
|
| Buffer: |
0.2 M glycine-HCl, 80 mM NaCl, pH: 2.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 29
|
Dependence of the Nanoscale Composite Morphology of Fe3O4 Nanoparticle-Infused Lysozyme Amyloid Fibrils on Timing of Infusion: A Combined SAXS and AFM Study
Molecules 26(16):4864 (2021)
Schroer M, Hu P, Tomasovicova N, Batkova M, Zakutanska K, Wu P, Kopcansky P
|
| RgGuinier |
22.5 |
nm |
| Dmax |
95.0 |
nm |
|
|
UniProt ID: P00698 (None-None) lysozyme amyloid fibril
UniProt ID: None (None-None) Fe3O4 nanoparticles; nominal diameter 20 nm (hydrodynamic diameter)
|
|
|
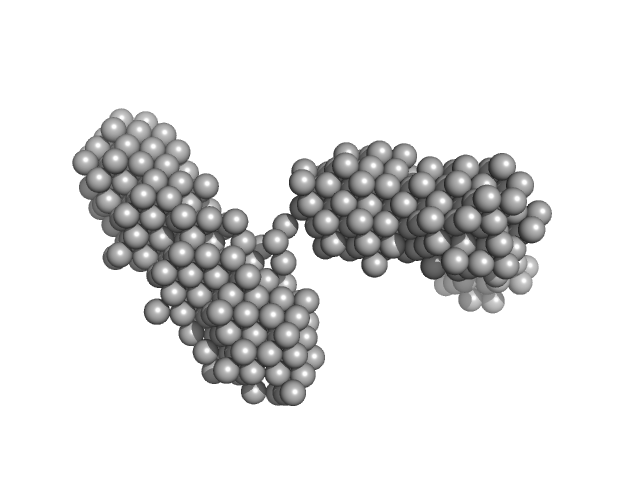
|
| Sample: |
Lysozyme amyloid fibril, 1 kDa Gallus gallus protein
Fe3O4 nanoparticles; nominal diameter 20 nm (hydrodynamic diameter) monomer, 1 kDa
|
| Buffer: |
0.2 M glycine-HCl, 80 mM NaCl, pH: 2.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 29
|
Dependence of the Nanoscale Composite Morphology of Fe3O4 Nanoparticle-Infused Lysozyme Amyloid Fibrils on Timing of Infusion: A Combined SAXS and AFM Study
Molecules 26(16):4864 (2021)
Schroer M, Hu P, Tomasovicova N, Batkova M, Zakutanska K, Wu P, Kopcansky P
|
| RgGuinier |
23.6 |
nm |
| Dmax |
90.0 |
nm |
|
|
UniProt ID: P00698 (None-None) lysozyme amyloid fibril
UniProt ID: None (None-None) Fe3O4 nanoparticles; nominal diameter 20 nm (hydrodynamic diameter)
|
|
|
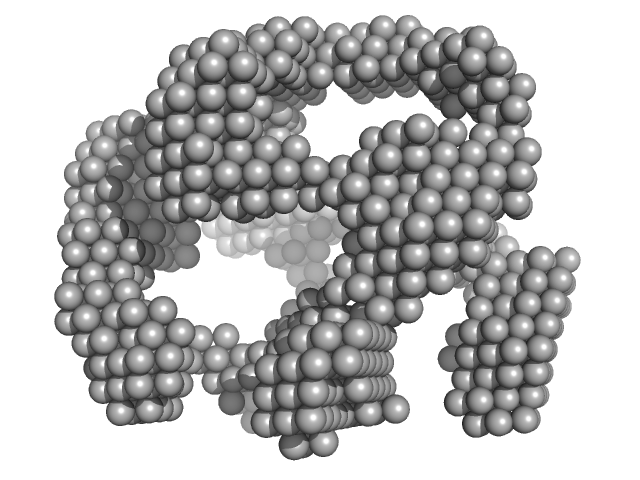
|
| Sample: |
Lysozyme amyloid fibril, 1 kDa Gallus gallus protein
Fe3O4 nanoparticles; nominal diameter 20 nm (hydrodynamic diameter) monomer, 1 kDa
|
| Buffer: |
0.2 M glycine-HCl, 80 mM NaCl, pH: 2.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Sep 2
|
Dependence of the Nanoscale Composite Morphology of Fe3O4 Nanoparticle-Infused Lysozyme Amyloid Fibrils on Timing of Infusion: A Combined SAXS and AFM Study
Molecules 26(16):4864 (2021)
Schroer M, Hu P, Tomasovicova N, Batkova M, Zakutanska K, Wu P, Kopcansky P
|
| RgGuinier |
31.0 |
nm |
| Dmax |
75.0 |
nm |
|
|
UniProt ID: Q8IAR7 (1-373) Tyrosine--tRNA ligase
|
|
|
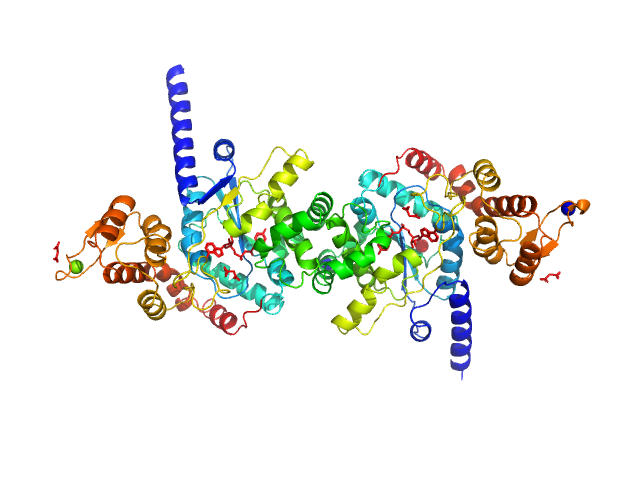
|
| Sample: |
Tyrosine--tRNA ligase dimer, 87 kDa Plasmodium falciparum (isolate … protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 0.5 mM TCEP, 0.1% sodium azide, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2021 Jun 4
|
Reaction hijacking of tyrosine tRNA synthetase as a new whole-of-life-cycle antimalarial strategy
Science 376(6597):1074-1079 (2022)
Xie S, Metcalfe R, Dunn E, Morton C, Huang S, Puhalovich T, Du Y, Wittlin S, Nie S, Luth M, Ma L, Kim M, Pasaje C, Kumpornsin K, Giannangelo C, Houghton F, Churchyard A, Famodimu M, Barry D, Gillett D, Dey S, Kosasih C, Newman W, Niles J, Lee M, Baum J, Ottilie S, Winzeler E, Creek D, Williamson N, Parker M, Brand S, Langston S, Dick L, Griffin M, Gould A, Tilley L
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
126 |
nm3 |
|
|
UniProt ID: Q8IAR7 (2-373) Tyrosine--tRNA ligase (S234C)
|
|
|
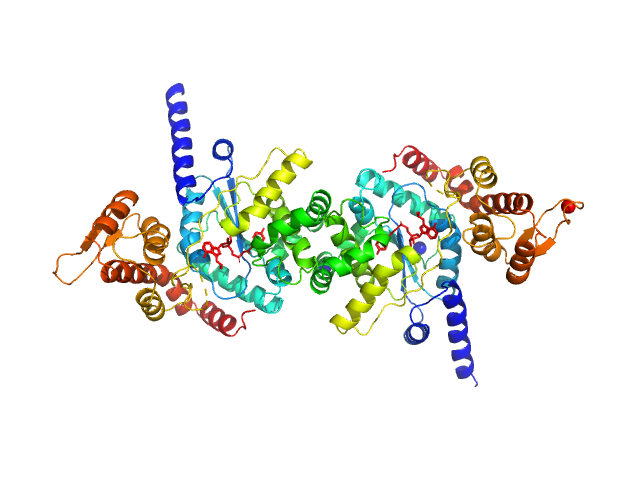
|
| Sample: |
Tyrosine--tRNA ligase (S234C) dimer, 87 kDa Plasmodium falciparum (isolate … protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 0.5 mM TCEP, 0.1% sodium azide, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2021 Jun 4
|
Reaction hijacking of tyrosine tRNA synthetase as a new whole-of-life-cycle antimalarial strategy
Science 376(6597):1074-1079 (2022)
Xie S, Metcalfe R, Dunn E, Morton C, Huang S, Puhalovich T, Du Y, Wittlin S, Nie S, Luth M, Ma L, Kim M, Pasaje C, Kumpornsin K, Giannangelo C, Houghton F, Churchyard A, Famodimu M, Barry D, Gillett D, Dey S, Kosasih C, Newman W, Niles J, Lee M, Baum J, Ottilie S, Winzeler E, Creek D, Williamson N, Parker M, Brand S, Langston S, Dick L, Griffin M, Gould A, Tilley L
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
124 |
nm3 |
|
|
UniProt ID: P54577 (1-364) Tyrosine--tRNA ligase, cytoplasmic
|
|
|
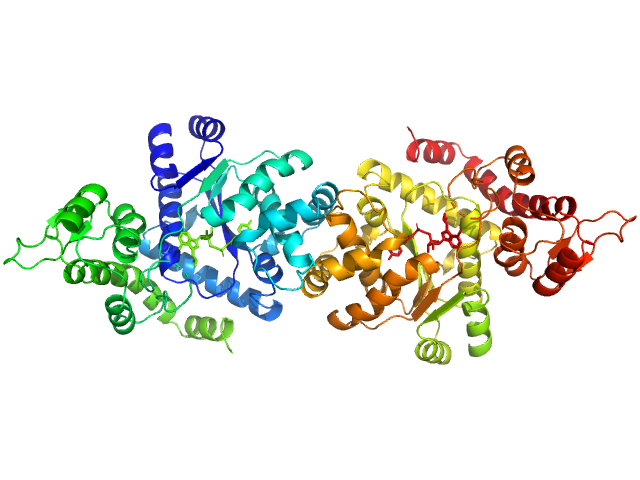
|
| Sample: |
Tyrosine--tRNA ligase, cytoplasmic dimer, 82 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 0.5 mM TCEP, 0.1% sodium azide, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2021 Jun 4
|
Reaction hijacking of tyrosine tRNA synthetase as a new whole-of-life-cycle antimalarial strategy
Science 376(6597):1074-1079 (2022)
Xie S, Metcalfe R, Dunn E, Morton C, Huang S, Puhalovich T, Du Y, Wittlin S, Nie S, Luth M, Ma L, Kim M, Pasaje C, Kumpornsin K, Giannangelo C, Houghton F, Churchyard A, Famodimu M, Barry D, Gillett D, Dey S, Kosasih C, Newman W, Niles J, Lee M, Baum J, Ottilie S, Winzeler E, Creek D, Williamson N, Parker M, Brand S, Langston S, Dick L, Griffin M, Gould A, Tilley L
|
| RgGuinier |
3.6 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
118 |
nm3 |
|
|