UniProt ID: P00698 (19-147) lysozyme amyloid fibril
|
|
|

|
| Sample: |
Lysozyme amyloid fibril, 14 kDa Gallus gallus protein
|
| Buffer: |
0.2 M glycine-HCl, 80 mM NaCl, pH: 2.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Sep 5
|
Effect of the concentration of protein and nanoparticles on the structure of biohybrid nanocomposites.
Biopolymers 111(2):e23342 (2020)
Majorošová J, Schroer MA, Tomašovičová N, Batková M, Hu PS, Kubovčíková M, Svergun DI, Kopčanský P
|
| RgGuinier |
30.2 |
nm |
| Dmax |
120.0 |
nm |
|
|
UniProt ID: P00698 (None-None) lysozyme amyloid fibril
UniProt ID: None (None-None) Fe3O4 nanoparticles; radius 5.6 nm (AFM based)
|
|
|

|
| Sample: |
Lysozyme amyloid fibril, 1 kDa Gallus gallus protein
Fe3O4 nanoparticles; radius 5.6 nm (AFM based) monomer, 1 kDa
|
| Buffer: |
0.2 M glycine-HCl, 80 mM NaCl, pH: 2.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Sep 8
|
Effect of the concentration of protein and nanoparticles on the structure of biohybrid nanocomposites.
Biopolymers 111(2):e23342 (2020)
Majorošová J, Schroer MA, Tomašovičová N, Batková M, Hu PS, Kubovčíková M, Svergun DI, Kopčanský P
|
| RgGuinier |
29.4 |
nm |
| Dmax |
80.0 |
nm |
|
|
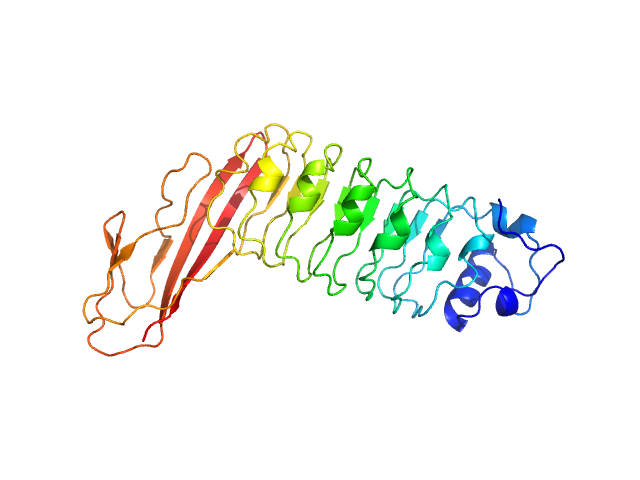
UniProt ID: P0DQD3 (1-320) Internalin B
|
|
|
|
| Sample: |
Internalin B monomer, 36 kDa Listeria monocytogenes serotype … protein
|
| Buffer: |
25mM Na-phosphate buffer, 150 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2003 Nov 11
|
X-ray and Neutron Small-Angle Scattering Analysis of the Complex Formed by the Met Receptor and the Listeria monocytogenes Invasion Protein InlB
Journal of Molecular Biology 377(2):489-500 (2008)
Niemann H, Petoukhov M, Härtlein M, Moulin M, Gherardi E, Timmins P, Heinz D, Svergun D
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
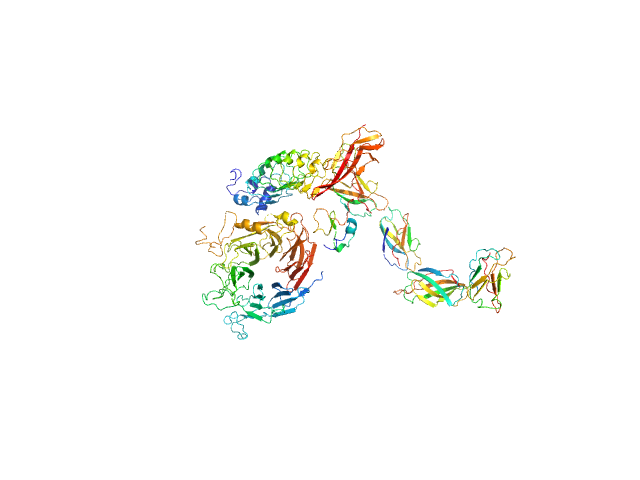
UniProt ID: P16056 (25-927) Hepatocyte growth factor receptor
UniProt ID: P0DQD3 (1-320) Internalin B
|
|
|
|
| Sample: |
Hepatocyte growth factor receptor monomer, 100 kDa Mus musculus protein
Internalin B monomer, 36 kDa Listeria monocytogenes serotype … protein
|
| Buffer: |
25mM Na-phosphate buffer, 150 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2004 Mar 1
|
X-ray and Neutron Small-Angle Scattering Analysis of the Complex Formed by the Met Receptor and the Listeria monocytogenes Invasion Protein InlB
Journal of Molecular Biology 377(2):489-500 (2008)
Niemann H, Petoukhov M, Härtlein M, Moulin M, Gherardi E, Timmins P, Heinz D, Svergun D
|
| RgGuinier |
4.6 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
270 |
nm3 |
|
|
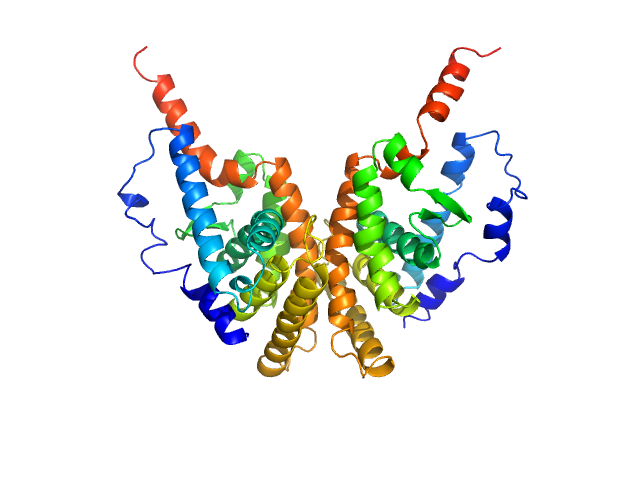
UniProt ID: P19793 (231-458) Retinoic acid receptor RXR-alpha
|
|
|
|
| Sample: |
Retinoic acid receptor RXR-alpha dimer, 51 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 100 mM KCl, 5% glycerol, 2 mM Chaps, and 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Oct 7
|
Solution Behavior of the Intrinsically Disordered N-Terminal Domain of Retinoid X Receptor α in the Context of the Full-Length Protein
Biochemistry 55(12):1741-1748 (2016)
Belorusova A, Osz J, Petoukhov M, Peluso-Iltis C, Kieffer B, Svergun D, Rochel N
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
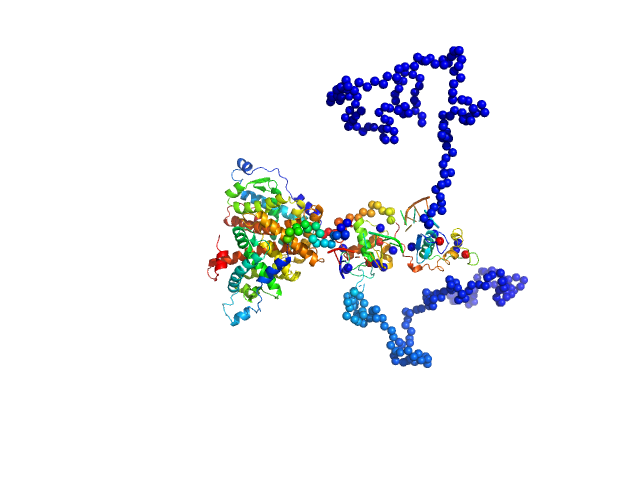
UniProt ID: P19793 (1-462) Retinoic acid receptor RXR-alpha
UniProt ID: None (None-None) Ramp2 DNA
|
|
|
|
| Sample: |
Retinoic acid receptor RXR-alpha dimer, 102 kDa Homo sapiens protein
Ramp2 DNA monomer, 11 kDa DNA
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 50 mM KCl, 5% glycerol, 2 mM Chaps, and 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 May 7
|
Solution Behavior of the Intrinsically Disordered N-Terminal Domain of Retinoid X Receptor α in the Context of the Full-Length Protein
Biochemistry 55(12):1741-1748 (2016)
Belorusova A, Osz J, Petoukhov M, Peluso-Iltis C, Kieffer B, Svergun D, Rochel N
|
| RgGuinier |
4.4 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
161 |
nm3 |
|
|
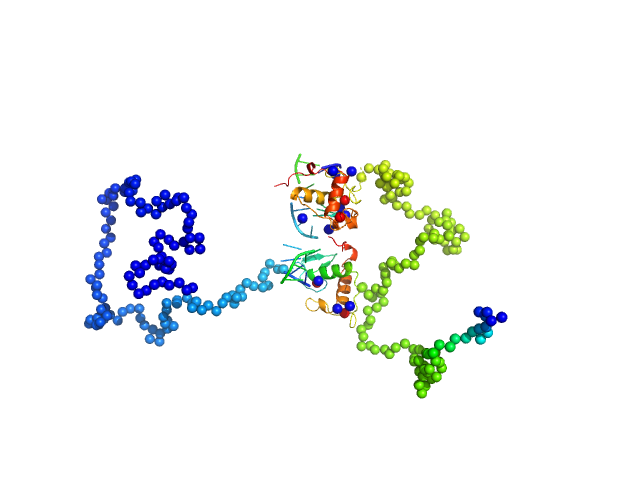
UniProt ID: None (None-None) Ramp2 DNA
UniProt ID: P19793 (1-206) Retinoic acid receptor RXR-alpha
|
|
|
|
| Sample: |
Ramp2 DNA monomer, 11 kDa DNA
Retinoic acid receptor RXR-alpha dimer, 44 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 50 mM KCl, 5% glycerol, 2 mM Chaps, and 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Jul 17
|
Solution Behavior of the Intrinsically Disordered N-Terminal Domain of Retinoid X Receptor α in the Context of the Full-Length Protein
Biochemistry 55(12):1741-1748 (2016)
Belorusova A, Osz J, Petoukhov M, Peluso-Iltis C, Kieffer B, Svergun D, Rochel N
|
| RgGuinier |
4.3 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
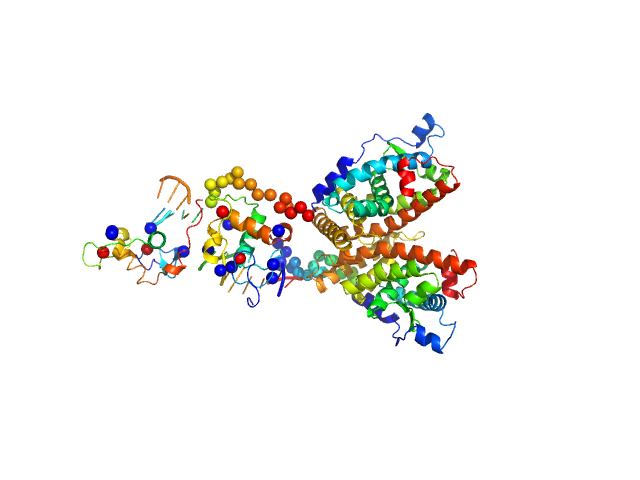
UniProt ID: None (None-None) Ramp2 DNA
UniProt ID: P19793 (None-None) Retinoic acid receptor RXR-alpha
|
|
|
|
| Sample: |
Ramp2 DNA monomer, 11 kDa DNA
Retinoic acid receptor RXR-alpha dimer, 74 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 50 mM KCl, 5% glycerol, 2 mM Chaps, and 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Oct 7
|
Solution Behavior of the Intrinsically Disordered N-Terminal Domain of Retinoid X Receptor α in the Context of the Full-Length Protein
Biochemistry 55(12):1741-1748 (2016)
Belorusova A, Osz J, Petoukhov M, Peluso-Iltis C, Kieffer B, Svergun D, Rochel N
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.2 |
nm |
|
|
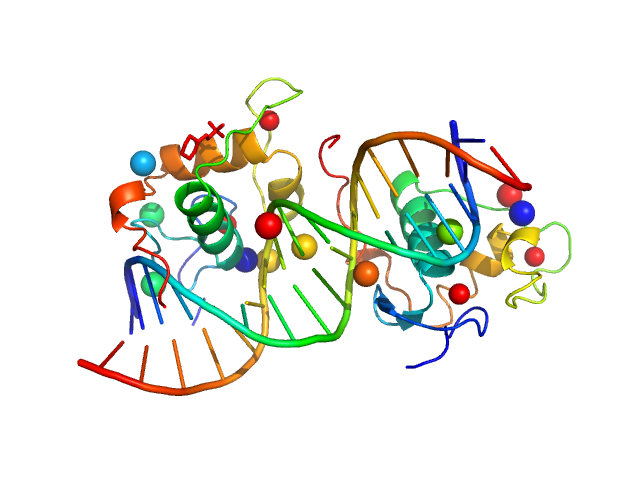
UniProt ID: None (None-None) Ramp2 DNA
UniProt ID: P19793 (None-None) Retinoic acid receptor RXR-alpha
|
|
|
|
| Sample: |
Ramp2 DNA monomer, 11 kDa DNA
Retinoic acid receptor RXR-alpha dimer, 20 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 50 mM KCl, 5% glycerol, 2 mM Chaps, and 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Oct 7
|
Solution Behavior of the Intrinsically Disordered N-Terminal Domain of Retinoid X Receptor α in the Context of the Full-Length Protein
Biochemistry 55(12):1741-1748 (2016)
Belorusova A, Osz J, Petoukhov M, Peluso-Iltis C, Kieffer B, Svergun D, Rochel N
|
| RgGuinier |
1.9 |
nm |
| Dmax |
5.8 |
nm |
|
|
UniProt ID: P62508 (122-458) Estrogen-related receptor gamma
UniProt ID: None (None-None) Inverse repeat IR3 DNA
|
|
|

|
| Sample: |
Estrogen-related receptor gamma hexamer, 230 kDa Homo sapiens protein
Inverse repeat IR3 DNA trimer, 46 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 100 mM KCl, 5 mM MgCl2, 1% (v/v) glycerol, and 1 mM CHAPS, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Mar 3
|
Reconstruction of Quaternary Structure from X-ray Scattering by Equilibrium Mixtures of Biological Macromolecules
Biochemistry 52(39):6844-6855 (2013)
Petoukhov M, Billas I, Takacs M, Graewert M, Moras D, Svergun D
|
| RgGuinier |
4.7 |
nm |
| Dmax |
14.6 |
nm |
|
|