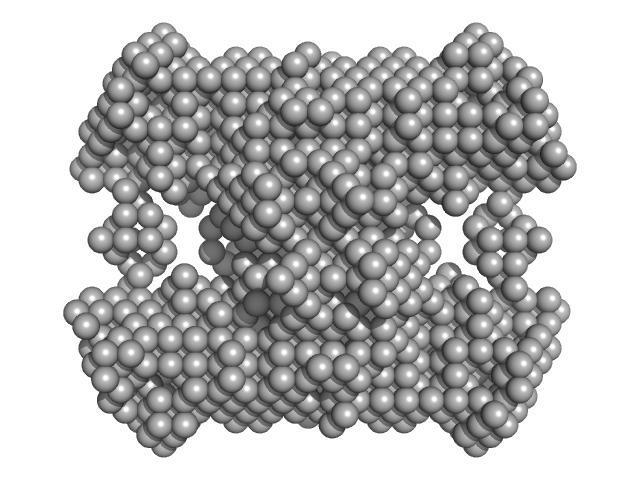
UniProt ID: Q59RW5 (1-493) N-acetylglucosamine kinase 1
|
|
|
|
| Sample: |
N-acetylglucosamine kinase 1 tetramer, 219 kDa Candida albicans (strain … protein
|
| Buffer: |
0.2 M magnesium acetate, 0.1 M sodium cacodylate, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 Apr 28
|
The crystal and solution studies of glucosamine-6-phosphate synthase from Candida albicans.
J Mol Biol 372(3):672-88 (2007)
Raczynska J, Olchowy J, Konariev PV, Svergun DI, Milewski S, Rypniewski W
|
| RgGuinier |
5.1 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
421 |
nm3 |
|
|
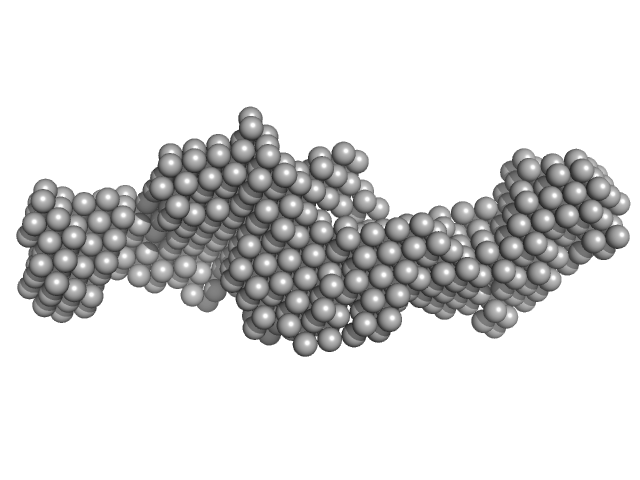
UniProt ID: Q7Z417 (1-540) Nuclear fragile X mental retardation-interacting protein 2
|
|
|
|
| Sample: |
Nuclear fragile X mental retardation-interacting protein 2 monomer, 59 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl pH 8.0, 5 mM TCEP, pH: |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Oct 11
|
A study of the ultrastructure of fragile-X-related proteins.
Biochem J 419(2):347-57 (2009)
Sjekloća L, Konarev PV, Eccleston J, Taylor IA, Svergun DI, Pastore A
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
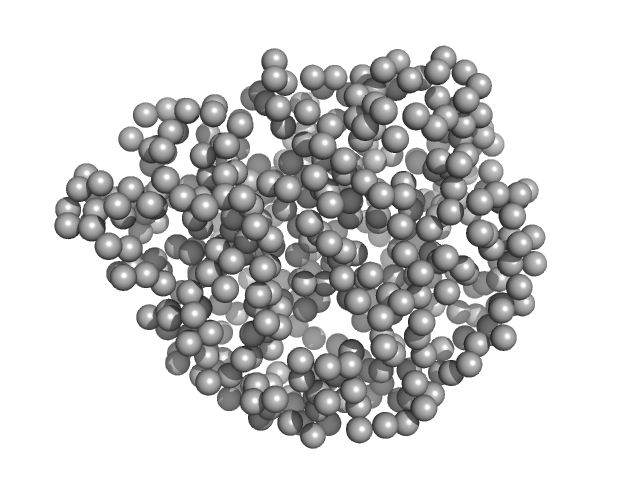
UniProt ID: K6THC9 (1-438) Lanthionine synthetase C-like protein
|
|
|
|
| Sample: |
Lanthionine synthetase C-like protein monomer, 50 kDa Clostridium sp. Maddingley … protein
|
| Buffer: |
50 mM HEPES, 500 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 Q-Xoom, Center for Structural Studies, Heinrich-Heine-University on 2019 Nov 28
|
The structure of MadC from Clostridium maddingley reveals new insights into class I lanthipeptide cyclases
Frontiers in Microbiology 13 (2023)
Knospe C, Kamel M, Spitz O, Hoeppner A, Galle S, Reiners J, Kedrov A, Smits S, Schmitt L
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
78 |
nm3 |
|
|
UniProt ID: Q9UBN7 (2-1215) Histone deacetylase 6
|
|
|
|
| Sample: |
Histone deacetylase 6 monomer, 131 kDa Homo sapiens protein
|
| Buffer: |
30 mM HEPES, 140 mM NaCl, 10 mM KCl, 0.25 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 10
|
In-solution structure and oligomerization of human histone deacetylase 6 - an integrative approach.
FEBS J (2022)
Shukla S, Komarek J, Novakova Z, Nedvedova J, Ustinova K, Vankova P, Kadek A, Uetrecht C, Mertens H, Barinka C
|
| RgGuinier |
7.0 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
316 |
nm3 |
|
|
UniProt ID: Q9HD67 (883-933) Unconventional myosin-X component
|
|
|
|
| Sample: |
Unconventional myosin-X component dimer, 15 kDa Homo sapiens protein
|
| Buffer: |
HEPES, 5% glycerol, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL-15A2, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2021 Mar 16
|
K
-edge anomalous SAXS for protein solution structure modeling
Acta Crystallographica Section D Structural Biology 78(2) (2022)
Virk K, Yonezawa K, Choukate K, Singh L, Shimizu N, Chaudhuri B
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
UniProt ID: Q9HD67 (883-933) Unconventional myosin-X
|
|
|
|
| Sample: |
Unconventional myosin-X dimer, 15 kDa Homo sapiens protein
|
| Buffer: |
HEPES, 5% glycerol, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL-15A2, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2021 Mar 16
|
K
-edge anomalous SAXS for protein solution structure modeling
Acta Crystallographica Section D Structural Biology 78(2) (2022)
Virk K, Yonezawa K, Choukate K, Singh L, Shimizu N, Chaudhuri B
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
UniProt ID: Q9HD67 (883-933) Unconventional myosin-X component
|
|
|
|
| Sample: |
Unconventional myosin-X component dimer, 15 kDa Homo sapiens protein
|
| Buffer: |
HEPES, 5% glycerol, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL-15A2, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2021 Mar 16
|
K
-edge anomalous SAXS for protein solution structure modeling
Acta Crystallographica Section D Structural Biology 78(2) (2022)
Virk K, Yonezawa K, Choukate K, Singh L, Shimizu N, Chaudhuri B
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
UniProt ID: Q9HD67 (883-933) Unconventional myosin-X component
|
|
|
|
| Sample: |
Unconventional myosin-X component dimer, 15 kDa Homo sapiens protein
|
| Buffer: |
HEPES, 5% glycerol, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL-15A2, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2021 Mar 16
|
K
-edge anomalous SAXS for protein solution structure modeling
Acta Crystallographica Section D Structural Biology 78(2) (2022)
Virk K, Yonezawa K, Choukate K, Singh L, Shimizu N, Chaudhuri B
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
UniProt ID: Q9HD67 (883-933) Unconventional myosin-X component
|
|
|
|
| Sample: |
Unconventional myosin-X component dimer, 15 kDa Homo sapiens protein
|
| Buffer: |
HEPES, 5% glycerol, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL-15A2, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2021 Mar 16
|
K
-edge anomalous SAXS for protein solution structure modeling
Acta Crystallographica Section D Structural Biology 78(2) (2022)
Virk K, Yonezawa K, Choukate K, Singh L, Shimizu N, Chaudhuri B
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
UniProt ID: Q9HD67 (883-933) Unconventional myosin-X component
|
|
|
|
| Sample: |
Unconventional myosin-X component dimer, 15 kDa Homo sapiens protein
|
| Buffer: |
HEPES, 5% glycerol, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL-15A2, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2021 Mar 16
|
K
-edge anomalous SAXS for protein solution structure modeling
Acta Crystallographica Section D Structural Biology 78(2) (2022)
Virk K, Yonezawa K, Choukate K, Singh L, Shimizu N, Chaudhuri B
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
24 |
nm3 |
|
|