UniProt ID: P10537 (2-499) Beta-amylase
|
|
|
|
| Sample: |
Beta-amylase tetramer, 224 kDa Ipomoea batatas protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, and 0.2 mM TCEP, pH: 7.3 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Dec 7
|
The BAM7
gene in Zea mays
encodes a protein with similar structural and catalytic properties to Arabidopsis
BAM2
Acta Crystallographica Section D Structural Biology 78(5) (2022)
Ravenburg C, Riney M, Monroe J, Berndsen C
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
296 |
nm3 |
|
|
UniProt ID: Q8TDB6 (1-200) E3 ubiquitin-protein ligase DTX3L
|
|
|
|
| Sample: |
E3 ubiquitin-protein ligase DTX3L pentamer, 114 kDa Homo sapiens protein
|
| Buffer: |
30 mM HEPES, 350 mM NaCl, 10% glycerol, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Feb 6
|
Reconstitution of the DTX3L-PARP9 complex reveals determinants for high affinity heterodimerization and multimeric assembly.
Biochem J (2022)
Ashok Y, Vela-Rodríguez C, Yang CS, Alanen HI, Liu F, Paschal BM, Lehtiö L
|
| RgGuinier |
4.7 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
200 |
nm3 |
|
|
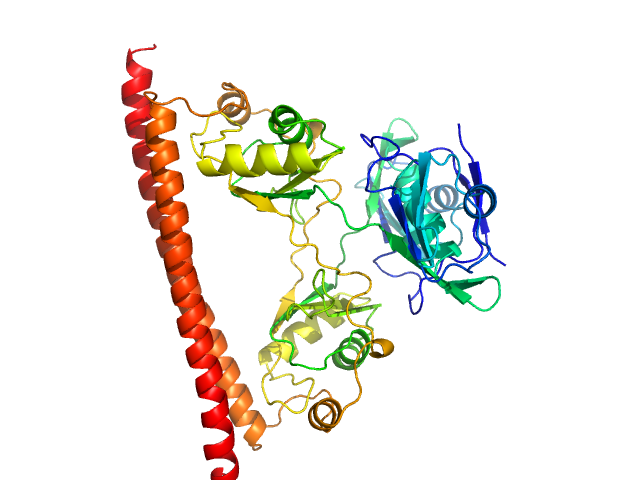
UniProt ID: Q15233 (53-312) Non-POU domain-containing octamer-binding protein
|
|
|
|
| Sample: |
Non-POU domain-containing octamer-binding protein dimer, 60 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-Cl (pH 7.5), 250 mM KCl, 50 mM L-proline, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Apr 28
|
Structural basis of dimerization and nucleic acid binding of human DBHS proteins NONO and PSPC1.
Nucleic Acids Res (2021)
Knott GJ, Chong YS, Passon DM, Liang XH, Deplazes E, Conte MR, Marshall AC, Lee M, Fox AH, Bond CS
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
96 |
nm3 |
|
|
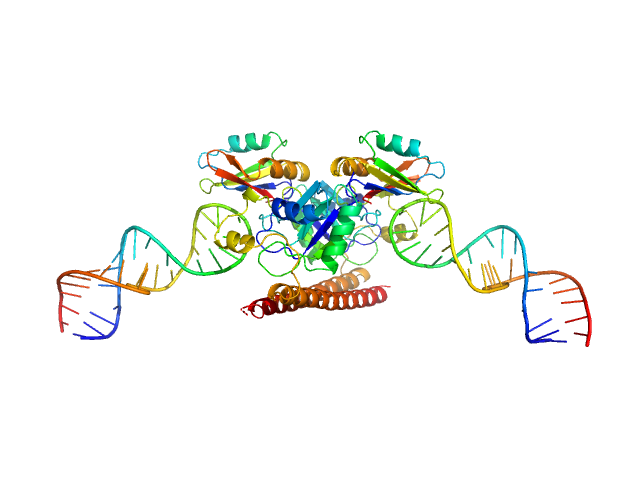
UniProt ID: Q15233 (53-312) Non-POU domain-containing octamer-binding protein
UniProt ID: None (None-None) 5-10-5 gapmer phosphorothioate antisense oligonucleotide tetramer
|
|
|
|
| Sample: |
Non-POU domain-containing octamer-binding protein dimer, 60 kDa Homo sapiens protein
5-10-5 gapmer phosphorothioate antisense oligonucleotide tetramer tetramer, 28 kDa
|
| Buffer: |
20 mM Tris-Cl (pH 7.5), 250 mM KCl, 50 mM L-proline, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Apr 28
|
Structural basis of dimerization and nucleic acid binding of human DBHS proteins NONO and PSPC1.
Nucleic Acids Res (2021)
Knott GJ, Chong YS, Passon DM, Liang XH, Deplazes E, Conte MR, Marshall AC, Lee M, Fox AH, Bond CS
|
| RgGuinier |
3.9 |
nm |
| Dmax |
18.4 |
nm |
| VolumePorod |
153 |
nm3 |
|
|
UniProt ID: Q9HC16 (1-384) DNA dC->dU-editing enzyme APOBEC-3G
|
|
|

|
| Sample: |
DNA dC->dU-editing enzyme APOBEC-3G tetramer, 186 kDa Homo sapiens protein
|
| Buffer: |
50 mM phosphate pH 6.0, 200 mM NaCl, 2 mM β-mercaptoethanol (β-ME), 5% glycerol, 200 µM Na2-EDTA, pH: 6 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Aug 6
|
Small-Angle X-ray Scattering (SAXS) Measurements of APOBEC3G Provide Structural Basis for Binding of Single-Stranded DNA and Processivity
Viruses 14(9):1974 (2022)
Barzak F, Ryan T, Mohammadzadeh N, Harjes S, Kvach M, Kurup H, Krause K, Chelico L, Filichev V, Harjes E, Jameson G
|
| RgGuinier |
4.2 |
nm |
| Dmax |
13.3 |
nm |
| VolumePorod |
350 |
nm3 |
|
|
UniProt ID: Q9HC16 (1-384) DNA dC->dU-editing enzyme APOBEC-3G
UniProt ID: None (None-None) 40-mer single stranded inhibitory DNA
|
|
|

|
| Sample: |
DNA dC->dU-editing enzyme APOBEC-3G tetramer, 186 kDa Homo sapiens protein
40-mer single stranded inhibitory DNA dimer, 24 kDa DNA
|
| Buffer: |
50 mM phosphate pH 6.0, 200 mM NaCl, 2 mM β-mercaptoethanol (β-ME), 5% glycerol, 200 µM Na2-EDTA, pH: 6 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Aug 6
|
Small-Angle X-ray Scattering (SAXS) Measurements of APOBEC3G Provide Structural Basis for Binding of Single-Stranded DNA and Processivity
Viruses 14(9):1974 (2022)
Barzak F, Ryan T, Mohammadzadeh N, Harjes S, Kvach M, Kurup H, Krause K, Chelico L, Filichev V, Harjes E, Jameson G
|
| RgGuinier |
4.7 |
nm |
| Dmax |
16.2 |
nm |
| VolumePorod |
395 |
nm3 |
|
|
UniProt ID: None (None-None) 40-mer single stranded inhibitory DNA
UniProt ID: Q9HC16 (1-384) DNA dC->dU-editing enzyme APOBEC-3G
|
|
|

|
| Sample: |
40-mer single stranded inhibitory DNA monomer, 12 kDa DNA
DNA dC->dU-editing enzyme APOBEC-3G monomer, 46 kDa Homo sapiens protein
|
| Buffer: |
50 mM phosphate pH 6.0, 200 mM NaCl, 2 mM β-mercaptoethanol (β-ME), 5% glycerol, 200 µM Na2-EDTA, pH: 6 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Aug 6
|
Small-Angle X-ray Scattering (SAXS) Measurements of APOBEC3G Provide Structural Basis for Binding of Single-Stranded DNA and Processivity
Viruses 14(9):1974 (2022)
Barzak F, Ryan T, Mohammadzadeh N, Harjes S, Kvach M, Kurup H, Krause K, Chelico L, Filichev V, Harjes E, Jameson G
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
118 |
nm3 |
|
|
UniProt ID: P08715 (1-1024) Hemolysin, plasmid (Hemolysin A)
|
|
|

|
| Sample: |
Hemolysin, plasmid (Hemolysin A) dimer, 220 kDa Escherichia coli UTI89 protein
|
| Buffer: |
100 mM HEPES pH 8.0, 250 mM NaCl, 10 mM CaCl2, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Mar 6
|
Identity Determinants of the Translocation Signal for a Type 1 Secretion System
Frontiers in Physiology 12 (2022)
Spitz O, Erenburg I, Kanonenberg K, Peherstorfer S, Lenders M, Reiners J, Ma M, Luisi B, Smits S, Schmitt L
|
| RgGuinier |
6.7 |
nm |
| Dmax |
25.3 |
nm |
| VolumePorod |
346 |
nm3 |
|
|
UniProt ID: Q9UKA9 (1-531) Polypyrimidine tract-binding protein 2
|
|
|

|
| Sample: |
Polypyrimidine tract-binding protein 2 monomer, 57 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 250 mM NaCl, 2 mM DTT, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2004 Feb 11
|
Structure and RNA interactions of the N-terminal RRM domains of PTB.
Structure 12(9):1631-43 (2004)
Simpson PJ, Monie TP, Szendröi A, Davydova N, Tyzack JK, Conte MR, Read CM, Cary PD, Svergun DI, Konarev PV, Curry S, Matthews S
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
UniProt ID: P04156 (1-253) Major prion protein
|
|
|

|
| Sample: |
Major prion protein 24-mer, 664 kDa Homo sapiens protein
|
| Buffer: |
5 mM sodium acetate, pH: 5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 Jun 2
|
Structural characterization of beta-sheeted oligomers formed on the pathway of oxidative prion protein aggregation in vitro.
J Struct Biol 157(2):308-20 (2007)
Redecke L, von Bergen M, Clos J, Konarev PV, Svergun DI, Fittschen UE, Broekaert JA, Bruns O, Georgieva D, Mandelkow E, Genov N, Betzel C
|
| RgGuinier |
9.8 |
nm |
| Dmax |
32.0 |
nm |
| VolumePorod |
3320 |
nm3 |
|
|