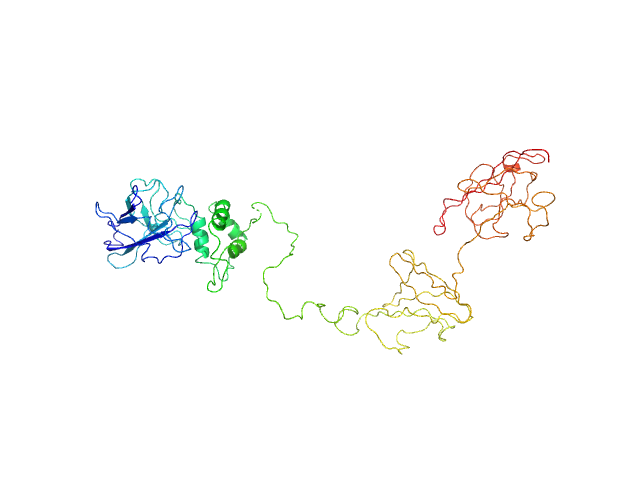
UniProt ID: A0A2N0URA4 (27-559) Dockerin domain-containing protein
|
|
|
|
| Sample: |
Dockerin domain-containing protein monomer, 57 kDa Ruminococcus bromii protein
|
| Buffer: |
phosphate buffered saline, 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Mar 28
|
Sas20 is a highly flexible starch-binding protein in the Ruminococcus bromii cell-surface amylosome
Journal of Biological Chemistry :101896 (2022)
Cerqueira F, Photenhauer A, Doden H, Brown A, Abdel-Hamid A, Moraïs S, Bayer E, Wawrzak Z, Cann I, Ridlon J, Hopkins J, Koropatkin N
|
| RgGuinier |
5.4 |
nm |
| Dmax |
20.3 |
nm |
| VolumePorod |
93 |
nm3 |
|
|
UniProt ID: A0A2N0URA4 (32-269) Dockerin domain-containing protein
|
|
|
|
| Sample: |
Dockerin domain-containing protein monomer, 26 kDa Ruminococcus bromii protein
|
| Buffer: |
phosphate buffered saline, 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Nov 14
|
Sas20 is a highly flexible starch-binding protein in the Ruminococcus bromii cell-surface amylosome
Journal of Biological Chemistry :101896 (2022)
Cerqueira F, Photenhauer A, Doden H, Brown A, Abdel-Hamid A, Moraïs S, Bayer E, Wawrzak Z, Cann I, Ridlon J, Hopkins J, Koropatkin N
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
45 |
nm3 |
|
|
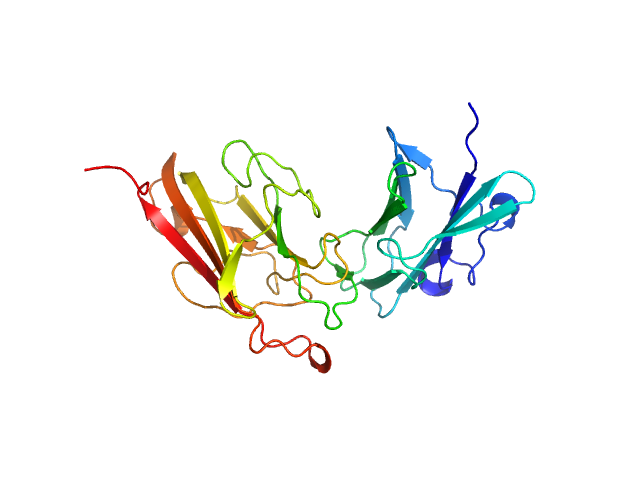
UniProt ID: A0A2N0URA4 (32-269) Dockerin domain-containing protein
|
|
|
|
| Sample: |
Dockerin domain-containing protein monomer, 26 kDa Ruminococcus bromii protein
|
| Buffer: |
phosphate buffered saline, 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Nov 14
|
Sas20 is a highly flexible starch-binding protein in the Ruminococcus bromii cell-surface amylosome
Journal of Biological Chemistry :101896 (2022)
Cerqueira F, Photenhauer A, Doden H, Brown A, Abdel-Hamid A, Moraïs S, Bayer E, Wawrzak Z, Cann I, Ridlon J, Hopkins J, Koropatkin N
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
UniProt ID: A0A2N0URA4 (32-269) Dockerin domain-containing protein
|
|
|
|
| Sample: |
Dockerin domain-containing protein monomer, 26 kDa Ruminococcus bromii protein
|
| Buffer: |
phosphate buffered saline, 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Nov 14
|
Sas20 is a highly flexible starch-binding protein in the Ruminococcus bromii cell-surface amylosome
Journal of Biological Chemistry :101896 (2022)
Cerqueira F, Photenhauer A, Doden H, Brown A, Abdel-Hamid A, Moraïs S, Bayer E, Wawrzak Z, Cann I, Ridlon J, Hopkins J, Koropatkin N
|
| RgGuinier |
5.2 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
UniProt ID: P00138 (1-127) Cytochrome c'
|
|
|
|
| Sample: |
Cytochrome c' monomer, 14 kDa Achromobacter xylosoxidans protein
|
| Buffer: |
Phosphate Buffer pD 1.7, pH: 1.7 |
| Experiment: |
SANS
data collected at KWS1, FRM2 on 2017 Aug 12
|
Open-Bundle Structure as the Unfolding Intermediate of Cytochrome c′ Revealed by Small Angle Neutron Scattering
Biomolecules 12(1):95 (2022)
Yamaguchi T, Akao K, Koutsioubas A, Frielinghaus H, Kohzuma T
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
13 |
nm3 |
|
|
UniProt ID: P00138 (1-127) Cytochrome c'
|
|
|
|
| Sample: |
Cytochrome c' dimer, 27 kDa Alcaligenes protein
|
| Buffer: |
Phosphate Buffer pD 6.4, pH: 6.4 |
| Experiment: |
SANS
data collected at KWS1, FRM2 on 2017 Aug 12
|
Open-Bundle Structure as the Unfolding Intermediate of Cytochrome c′ Revealed by Small Angle Neutron Scattering
Biomolecules 12(1):95 (2022)
Yamaguchi T, Akao K, Koutsioubas A, Frielinghaus H, Kohzuma T
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.5 |
nm |
| VolumePorod |
11 |
nm3 |
|
|
UniProt ID: P00138 (1-127) Cytochrome c'
|
|
|
|
| Sample: |
Cytochrome c' dimer, 27 kDa Alcaligenes protein
|
| Buffer: |
Phosphate Buffer pD 9.6, pH: 9.6 |
| Experiment: |
SANS
data collected at KWS1, FRM2 on 2017 Aug 12
|
Open-Bundle Structure as the Unfolding Intermediate of Cytochrome c′ Revealed by Small Angle Neutron Scattering
Biomolecules 12(1):95 (2022)
Yamaguchi T, Akao K, Koutsioubas A, Frielinghaus H, Kohzuma T
|
| RgGuinier |
1.9 |
nm |
| Dmax |
5.3 |
nm |
| VolumePorod |
10 |
nm3 |
|
|
UniProt ID: P00138 (1-127) Cytochrome c'
|
|
|
|
| Sample: |
Cytochrome c' monomer, 14 kDa Achromobacter xylosoxidans protein
|
| Buffer: |
Phosphate Buffer pD 13, pH: 13 |
| Experiment: |
SANS
data collected at KWS1, FRM2 on 2017 Aug 12
|
Open-Bundle Structure as the Unfolding Intermediate of Cytochrome c′ Revealed by Small Angle Neutron Scattering
Biomolecules 12(1):95 (2022)
Yamaguchi T, Akao K, Koutsioubas A, Frielinghaus H, Kohzuma T
|
| RgGuinier |
4.8 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
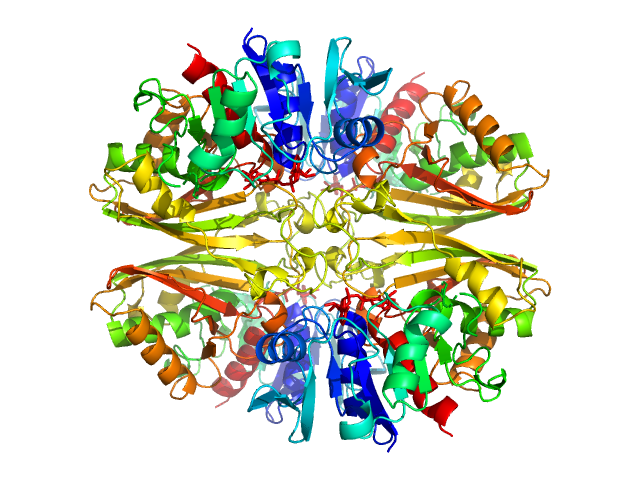
UniProt ID: P84998 (None-None) Glyceraldehyde-3-phosphate dehydrogenase 1
|
|
|
|
| Sample: |
Glyceraldehyde-3-phosphate dehydrogenase 1 tetramer, 142 kDa Kluyveromyces marxianus protein
|
| Buffer: |
150 mM NaCl, 1 mM beta-mercaptoethanol, 1 mM EDTA, 10 mM TrisHCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Mar 27
|
The Crystal and Solution Structures of Glyceraldehyde-3-phosphate Dehydrogenase Reveal Different Quaternary Structures
Journal of Biological Chemistry 281(44):33433-33440 (2006)
Ferreira-da-Silva F, Pereira P, Gales L, Roessle M, Svergun D, Moradas-Ferreira P, Damas A
|
| RgGuinier |
4.2 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
234 |
nm3 |
|
|
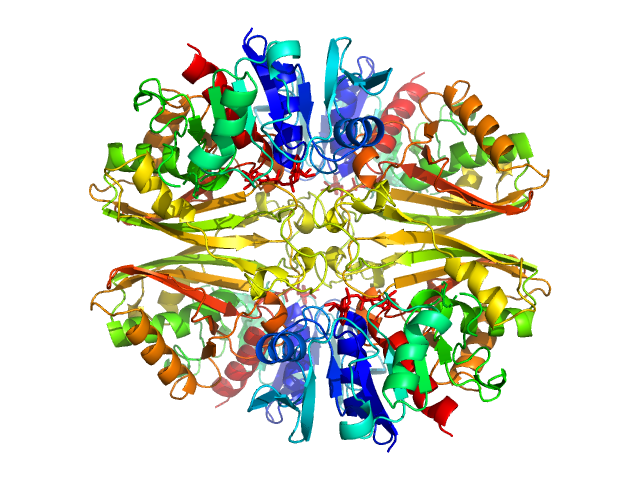
UniProt ID: P84998 (None-None) Glyceraldehyde-3-phosphate dehydrogenase 1 bound to NAD+
|
|
|
|
| Sample: |
Glyceraldehyde-3-phosphate dehydrogenase 1 bound to NAD+ tetramer, 142 kDa Kluyveromyces marxianus protein
|
| Buffer: |
150 mM NaCl, 1 mM beta-mercaptoethanol, 1 mM EDTA, 10 mM TrisHCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Mar 27
|
The Crystal and Solution Structures of Glyceraldehyde-3-phosphate Dehydrogenase Reveal Different Quaternary Structures
Journal of Biological Chemistry 281(44):33433-33440 (2006)
Ferreira-da-Silva F, Pereira P, Gales L, Roessle M, Svergun D, Moradas-Ferreira P, Damas A
|
| RgGuinier |
3.7 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
202 |
nm3 |
|
|