UniProt ID: P52003 (5-139) Multidrug resistance operon repressor
UniProt ID: None (None-None) 34 base pair double-stranded DNA
|
|
|
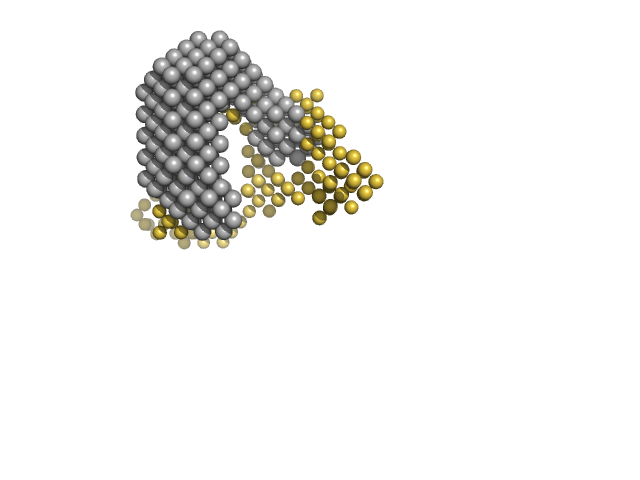
|
| Sample: |
Multidrug resistance operon repressor dimer, 32 kDa Pseudomonas aeruginosa protein
34 base pair double-stranded DNA monomer, 21 kDa synthetic construct DNA
|
| Buffer: |
20mM NaPO4, 150 mM NaCl, 10 mM DTT, pH: 7.1 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2018 May 30
|
Small-angle X-ray and neutron scattering of MexR and its complex with DNA supports a conformational selection binding model
Biophysical Journal (2022)
Caporaletti F, Pietras Z, Morad V, Mårtensson L, Gabel F, Wallner B, Martel A, Sunnerhagen M
|
| RgGuinier |
2.9 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
79 |
nm3 |
|
|
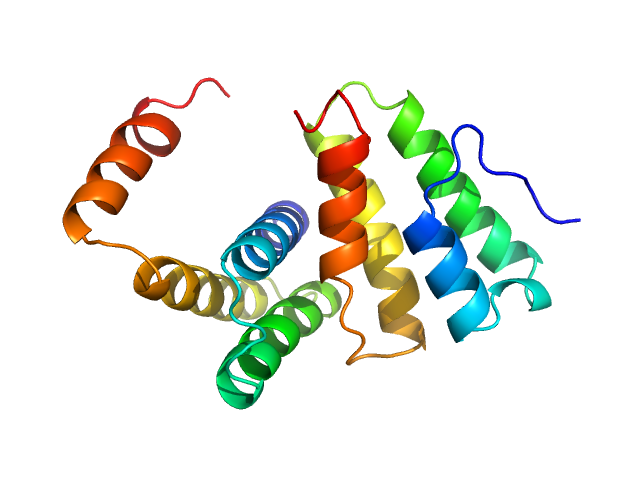
UniProt ID: P0C6U8 (3837-3919) Replicase polyprotein 1a
|
|
|
|
| Sample: |
Replicase polyprotein 1a dimer, 19 kDa Severe acute respiratory … protein
|
| Buffer: |
10 mM Tris-HCl, 200 mM NaCl, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Apr 2
|
Nonstructural Proteins 7 and 8 of Feline Coronavirus Form a 2:1 Heterotrimer That Exhibits Primer-Independent RNA Polymerase Activity
Journal of Virology 86(8):4444-4454 (2012)
Xiao Y, Ma Q, Restle T, Shang W, Svergun D, Ponnusamy R, Sczakiel G, Hilgenfeld R
|
|
|
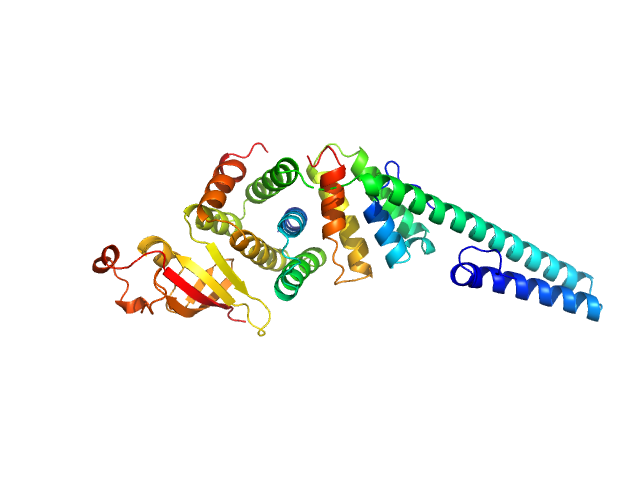
UniProt ID: P0C6U8 (3837-3919) Replicase polyprotein 1a
UniProt ID: P0C6U8 (3943-4140) Replicase polyprotein 1a
|
|
|
|
| Sample: |
Replicase polyprotein 1a dimer, 19 kDa Severe acute respiratory … protein
Replicase polyprotein 1a monomer, 22 kDa Severe acute respiratory … protein
|
| Buffer: |
10 mM Tris-HCl, 200 mM NaCl, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Apr 2
|
Nonstructural Proteins 7 and 8 of Feline Coronavirus Form a 2:1 Heterotrimer That Exhibits Primer-Independent RNA Polymerase Activity
Journal of Virology 86(8):4444-4454 (2012)
Xiao Y, Ma Q, Restle T, Shang W, Svergun D, Ponnusamy R, Sczakiel G, Hilgenfeld R
|
|
|
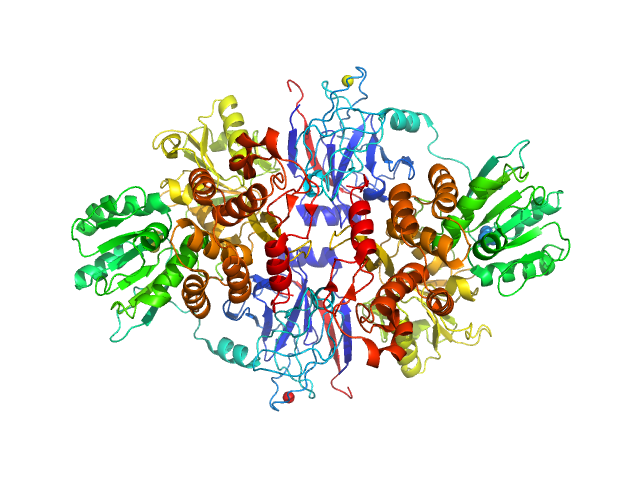
UniProt ID: P26663 (None-None) Genome polyprotein
|
|
|
|
| Sample: |
Genome polyprotein dimer, 137 kDa Hepatitis C virus … protein
|
| Buffer: |
25 mM Tris, 1 M NaCl, 10% glycerol, 1 mM TCEP, 0.1% β-octyl glucoside, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Dec 12
|
A macrocyclic HCV NS3/4A protease inhibitor interacts with protease and helicase residues in the complex with its full-length target
Proceedings of the National Academy of Sciences 108(52):21052-21056 (2011)
Schiering N, D'Arcy A, Villard F, Simic O, Kamke M, Monnet G, Hassiepen U, Svergun D, Pulfer R, Eder J, Raman P, Bodendorf U
|
|
|
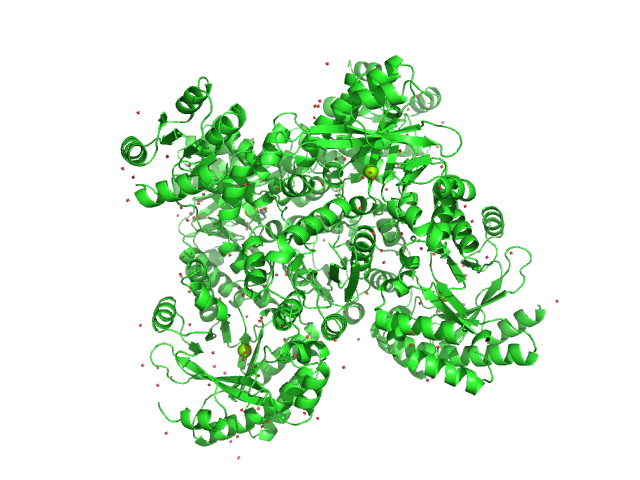
UniProt ID: Q9HX69 (None-None) RocR
|
|
|
|
| Sample: |
RocR tetramer, 171 kDa Pseudomonas aeruginosa (strain … protein
|
| Buffer: |
50 mM Tris–HCl, 250 mM NaCl, 10 mM imidazole, 5% glycerol, 0.5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Dec 9
|
Structural Insights into the Regulatory Mechanism of the Response Regulator RocR from Pseudomonas aeruginosa in Cyclic Di-GMP Signaling
Journal of Bacteriology 194(18):4837-4846 (2012)
Chen M, Kotaka M, Vonrhein C, Bricogne G, Rao F, Chuah M, Svergun D, Schneider G, Liang Z, Lescar J
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.0 |
nm |
|
|
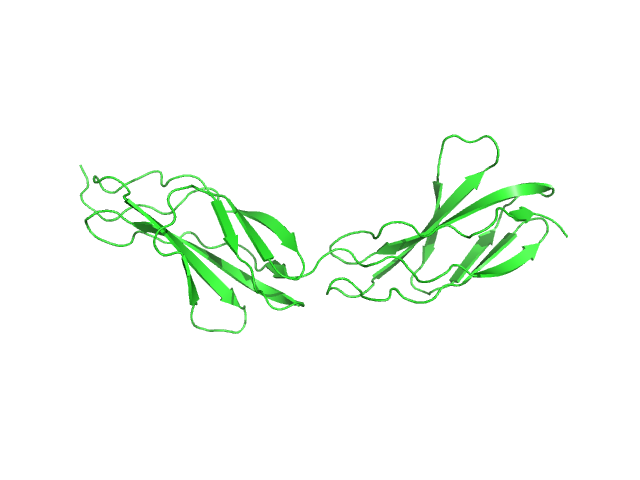
UniProt ID: Q8WZ42 (None-None) Titin
|
|
|
|
| Sample: |
Titin monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
100 mM NaCl, 50 mM Tris-HCl, 2mM DTT, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Jul 3
|
The Structure of the FnIII Tandem A77-A78 Points to a Periodically Conserved Architecture in the Myosin-Binding Region of Titin
Journal of Molecular Biology 401(5):843-853 (2010)
Bucher R, Svergun D, Muhle-Goll C, Mayans O
|
| RgGuinier |
2.5 |
nm |
| Dmax |
90.0 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
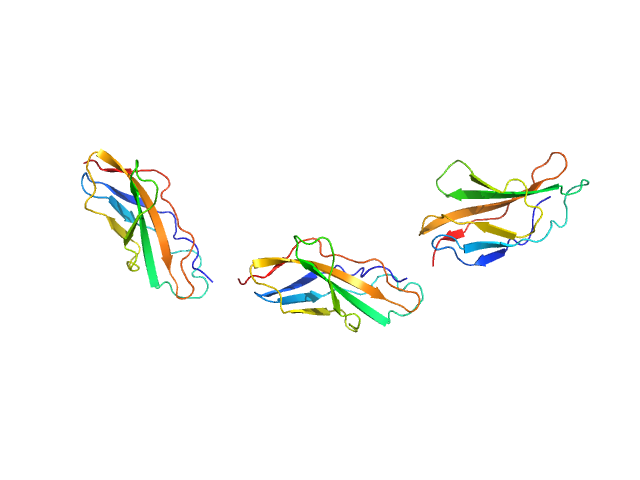
UniProt ID: Q8WZ42 (None-None) Titin
|
|
|
|
| Sample: |
Titin monomer, 32 kDa Homo sapiens protein
|
| Buffer: |
100 mM NaCl, 50 mM Tris-HCl, 2mM DTT, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Jul 3
|
The Structure of the FnIII Tandem A77-A78 Points to a Periodically Conserved Architecture in the Myosin-Binding Region of Titin
Journal of Molecular Biology 401(5):843-853 (2010)
Bucher R, Svergun D, Muhle-Goll C, Mayans O
|
| RgGuinier |
3.7 |
nm |
| Dmax |
130.0 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
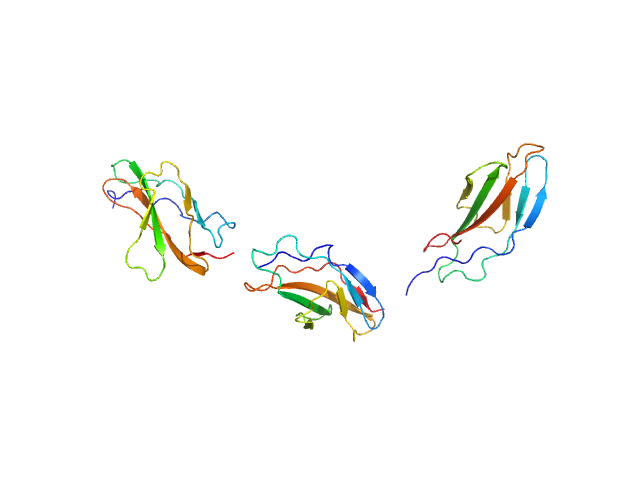
UniProt ID: Q8WZ42 (None-None) Titin
|
|
|
|
| Sample: |
Titin monomer, 32 kDa Homo sapiens protein
|
| Buffer: |
100 mM NaCl, 50 mM Tris-HCl, 2mM DTT, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Oct 6
|
The Structure of the FnIII Tandem A77-A78 Points to a Periodically Conserved Architecture in the Myosin-Binding Region of Titin
Journal of Molecular Biology 401(5):843-853 (2010)
Bucher R, Svergun D, Muhle-Goll C, Mayans O
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.0 |
nm |
|
|
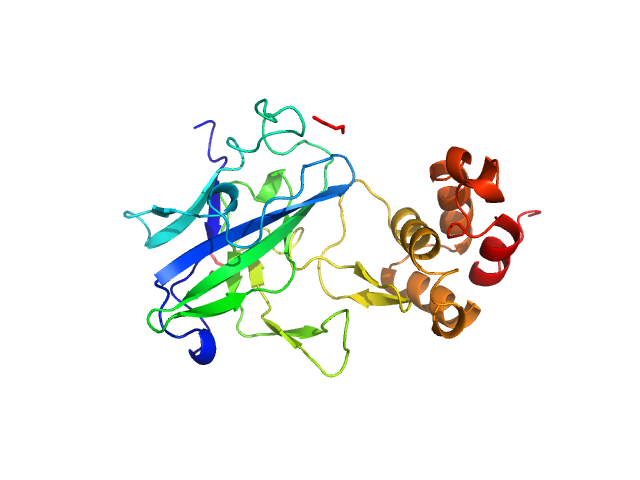
UniProt ID: A0A2N0URA4 (32-269) Dockerin domain-containing protein
|
|
|
|
| Sample: |
Dockerin domain-containing protein monomer, 26 kDa Ruminococcus bromii protein
|
| Buffer: |
phosphate buffered saline, 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Mar 28
|
Sas20 is a highly flexible starch-binding protein in the Ruminococcus bromii cell-surface amylosome
Journal of Biological Chemistry :101896 (2022)
Cerqueira F, Photenhauer A, Doden H, Brown A, Abdel-Hamid A, Moraïs S, Bayer E, Wawrzak Z, Cann I, Ridlon J, Hopkins J, Koropatkin N
|
| RgGuinier |
2.1 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
42 |
nm3 |
|
|
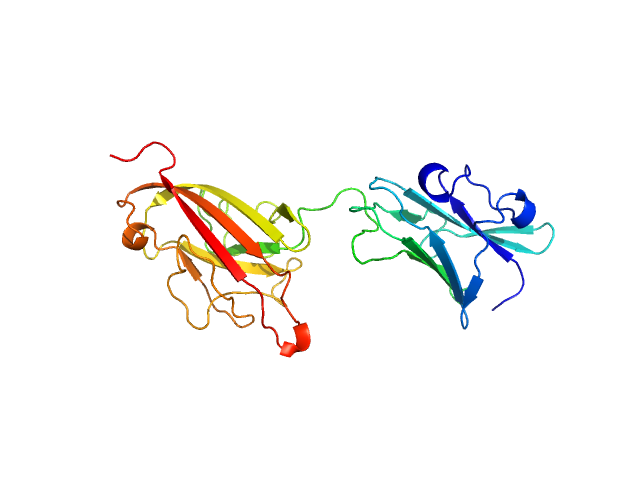
UniProt ID: A0A2N0URA4 (311-556) Dockerin domain-containing protein
|
|
|
|
| Sample: |
Dockerin domain-containing protein monomer, 26 kDa Ruminococcus bromii protein
|
| Buffer: |
phosphate buffered saline, 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Mar 28
|
Sas20 is a highly flexible starch-binding protein in the Ruminococcus bromii cell-surface amylosome
Journal of Biological Chemistry :101896 (2022)
Cerqueira F, Photenhauer A, Doden H, Brown A, Abdel-Hamid A, Moraïs S, Bayer E, Wawrzak Z, Cann I, Ridlon J, Hopkins J, Koropatkin N
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
35 |
nm3 |
|
|