UniProt ID: P23743 (1-197) Diacylglycerol kinase alpha
|
|
|

|
| Sample: |
Diacylglycerol kinase alpha monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
20 mM TrisHCl, 200 mM NaCl, 5 mM DTT, 5% v/v glycerol, 3 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Dec 8
|
Ca 2+
‐induced structural changes and intramolecular interactions in N‐terminal region of diacylglycerol kinase alpha
Protein Science 31(7) (2022)
Takahashi D, Yonezawa K, Okizaki Y, Caaveiro J, Ueda T, Shimada A, Sakane F, Shimizu N
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
UniProt ID: P23743 (1-197) Diacylglycerol kinase alpha
|
|
|

|
| Sample: |
Diacylglycerol kinase alpha monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
20mM TrisHCl, 200mM NaCl, 5mM DTT, 5% v/v Glycerol, 3mM EGTA, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Dec 8
|
Ca 2+
‐induced structural changes and intramolecular interactions in N‐terminal region of diacylglycerol kinase alpha
Protein Science 31(7) (2022)
Takahashi D, Yonezawa K, Okizaki Y, Caaveiro J, Ueda T, Shimada A, Sakane F, Shimizu N
|
| RgGuinier |
1.9 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
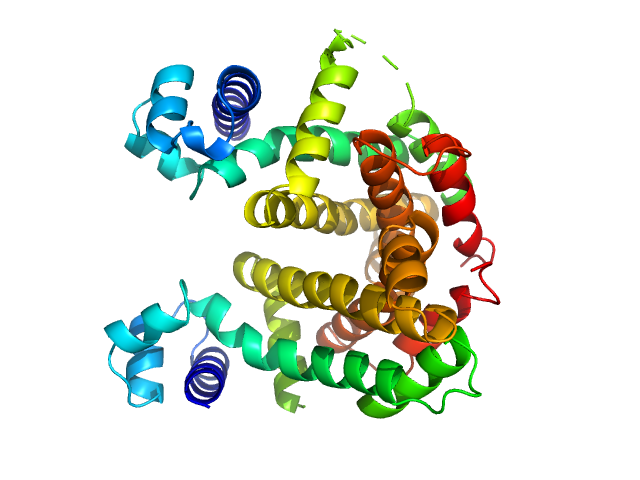
UniProt ID: Q9KYU4 (23-236) Putative tetR-family transcriptional regulator
|
|
|
|
| Sample: |
Putative tetR-family transcriptional regulator dimer, 47 kDa Streptomyces coelicolor (strain … protein
|
| Buffer: |
20 mM Tris/HCl, 400 mM NaCl, 50 mM imidazole, 1 mM DTT, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 May 2
|
Crystal Structures of Free and Ligand-Bound Forms of the TetR/AcrR-Like Regulator SCO3201 from Streptomyces coelicolor Suggest a Novel Allosteric Mechanism.
FEBS J (2022)
Werten S, Waack P, Palm GJ, Virolle MJ, Hinrichs W
|
| RgGuinier |
2.6 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
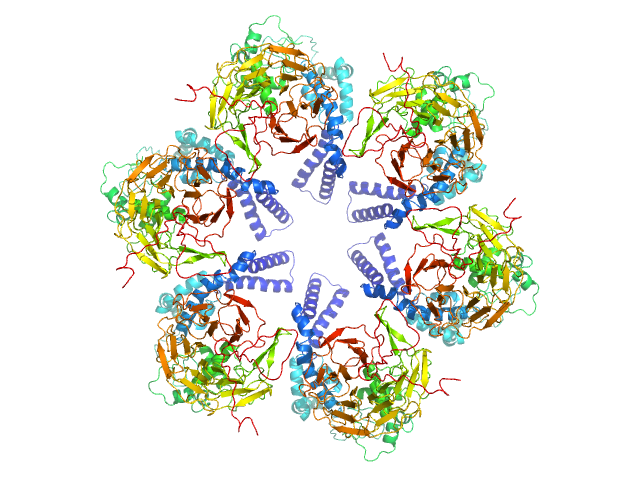
UniProt ID: Q8IDQ2 (189-726) Kelch protein K13
|
|
|
|
| Sample: |
Kelch protein K13 hexamer, 396 kDa Plasmodium falciparum (isolate … protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2020 Jul 25
|
Plasmodium falciparum Kelch13 and its artemisinin-resistant mutants assemble as hexamers in solution: a SAXS data driven modeling study.
FEBS J (2022)
Goel N, Dhiman K, Kalidas N, Mukhopadhyay A, Ashish F, Bhattacharjee S
|
| RgGuinier |
6.4 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
1630 |
nm3 |
|
|
UniProt ID: Q8IDQ2 (189-726) Kelch protein K13 (Truncated Kelch13-R539T, artemisinin-resistant mutation)
|
|
|

|
| Sample: |
Kelch protein K13 (Truncated Kelch13-R539T, artemisinin-resistant mutation) hexamer, 396 kDa Plasmodium falciparum (isolate … protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2020 Jul 25
|
Plasmodium falciparum Kelch13 and its artemisinin-resistant mutants assemble as hexamers in solution: a SAXS data driven modeling study.
FEBS J (2022)
Goel N, Dhiman K, Kalidas N, Mukhopadhyay A, Ashish F, Bhattacharjee S
|
| RgGuinier |
6.6 |
nm |
| Dmax |
20.5 |
nm |
| VolumePorod |
1600 |
nm3 |
|
|
UniProt ID: Q8IDQ2 (189-726) Kelch protein K13 (Truncated Kelch13-C580Y )
|
|
|

|
| Sample: |
Kelch protein K13 (Truncated Kelch13-C580Y ) hexamer, 397 kDa Plasmodium falciparum (isolate … protein
|
| Buffer: |
Phosphate Buffer Saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2020 Jul 28
|
Plasmodium falciparum Kelch13 and its artemisinin-resistant mutants assemble as hexamers in solution: a SAXS data driven modeling study.
FEBS J (2022)
Goel N, Dhiman K, Kalidas N, Mukhopadhyay A, Ashish F, Bhattacharjee S
|
| RgGuinier |
6.4 |
nm |
| Dmax |
18.4 |
nm |
| VolumePorod |
1800 |
nm3 |
|
|
UniProt ID: Q8IDQ2 (189-726) Kelch protein K13 (Truncated Kelch13-A578S)
|
|
|

|
| Sample: |
Kelch protein K13 (Truncated Kelch13-A578S) hexamer, 396 kDa Plasmodium falciparum (isolate … protein
|
| Buffer: |
Phosphate Buffer Saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2020 Jul 29
|
Plasmodium falciparum Kelch13 and its artemisinin-resistant mutants assemble as hexamers in solution: a SAXS data driven modeling study.
FEBS J (2022)
Goel N, Dhiman K, Kalidas N, Mukhopadhyay A, Ashish F, Bhattacharjee S
|
| RgGuinier |
6.1 |
nm |
| Dmax |
16.8 |
nm |
| VolumePorod |
1700 |
nm3 |
|
|
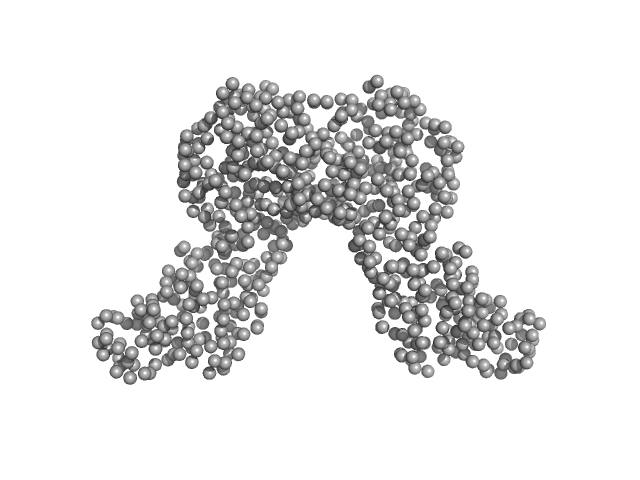
UniProt ID: A0A5P1UGY3 (1-361) Riboflavin biosynthesis protein RibD
|
|
|
|
| Sample: |
Riboflavin biosynthesis protein RibD dimer, 80 kDa Acinetobacter baumannii protein
|
| Buffer: |
150 mM NaCl, 10 mM Tris, 1 mM DTT, 5% v/v glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Dec 3
|
Recombinant AcnB, NrdR and RibD of Acinetobacter baumannii and their potential interaction with DNA adenine methyltransferase AamA.
Protein Expr Purif :106134 (2022)
Weber K, Doellinger J, Jeffries CM, Wilharm G
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
UniProt ID: V5VBJ4 (1-879) Aconitate hydratase B
|
|
|

|
| Sample: |
Aconitate hydratase B monomer, 96 kDa Acinetobacter baumannii protein
|
| Buffer: |
100 mM NaCl, 20 mM Tris, 1 mM DTT, 3% v/v glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Dec 3
|
Recombinant AcnB, NrdR and RibD of Acinetobacter baumannii and their potential interaction with DNA adenine methyltransferase AamA.
Protein Expr Purif :106134 (2022)
Weber K, Doellinger J, Jeffries CM, Wilharm G
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
150 |
nm3 |
|
|
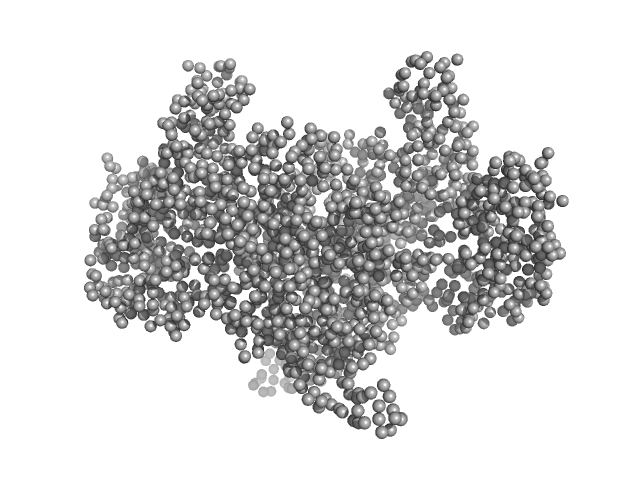
UniProt ID: V5VBJ4 (1-879) Aconitate hydratase B
|
|
|
|
| Sample: |
Aconitate hydratase B, 96 kDa Acinetobacter baumannii protein
|
| Buffer: |
100 mM NaCl, 20 mM Tris, 1 mM DTT, 3% v/v glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Dec 3
|
Recombinant AcnB, NrdR and RibD of Acinetobacter baumannii and their potential interaction with DNA adenine methyltransferase AamA.
Protein Expr Purif :106134 (2022)
Weber K, Doellinger J, Jeffries CM, Wilharm G
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
230 |
nm3 |
|
|