UniProt ID: A3M1A1 (1-152) Transcriptional repressor NrdR
|
|
|
|
| Sample: |
Transcriptional repressor NrdR, 19 kDa Acinetobacter baumannii (strain … protein
|
| Buffer: |
100 mM NaCl, 20 mM Tris, 1 mM DTT, 3% v/v glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Dec 3
|
Recombinant AcnB, NrdR and RibD of Acinetobacter baumannii and their potential interaction with DNA adenine methyltransferase AamA.
Protein Expr Purif :106134 (2022)
Weber K, Doellinger J, Jeffries CM, Wilharm G
|
| RgGuinier |
19.9 |
nm |
| Dmax |
92.6 |
nm |
|
|
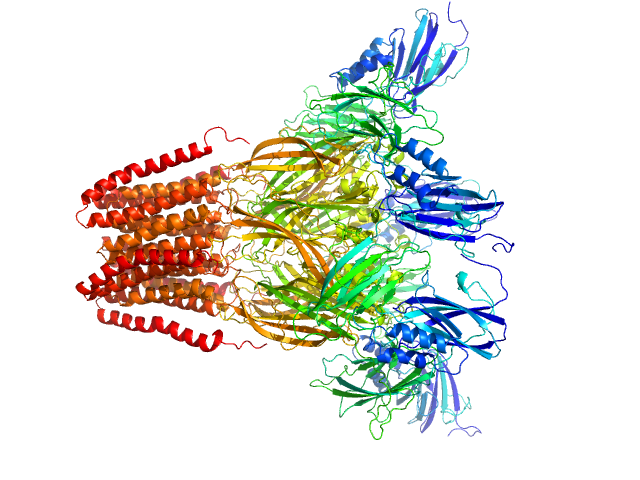
UniProt ID: V4JF97 (34-642) Neurotransmitter-gated ion-channel ligand-binding domain-containing protein
|
|
|
|
| Sample: |
Neurotransmitter-gated ion-channel ligand-binding domain-containing protein pentamer, 342 kDa Desulfofustis sp. PB-SRB1 protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 10 mM CaCl2, 0.5 mM deuterated n-Dodecyl-B-D-Maltoside, in D2O, pH: 7.5 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2020 Aug 23
|
Biophysical characterization of calcium-binding and modulatory-domain dynamics in a pentameric ligand-gated ion channel.
Proc Natl Acad Sci U S A 119(50):e2210669119 (2022)
Lycksell M, Rovšnik U, Hanke A, Martel A, Howard RJ, Lindahl E
|
| RgGuinier |
5.2 |
nm |
| Dmax |
16.8 |
nm |
| VolumePorod |
530 |
nm3 |
|
|
UniProt ID: V4JF97 (34-642) Neurotransmitter-gated ion-channel ligand-binding domain-containing protein
|
|
|
|
| Sample: |
Neurotransmitter-gated ion-channel ligand-binding domain-containing protein pentamer, 342 kDa Desulfofustis sp. PB-SRB1 protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 10 mM EDTA, 0.5 mM deuterated n-Dodecyl-B-D-Maltoside, in D2O, pH: 7.5 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2020 Aug 23
|
Biophysical characterization of calcium-binding and modulatory-domain dynamics in a pentameric ligand-gated ion channel.
Proc Natl Acad Sci U S A 119(50):e2210669119 (2022)
Lycksell M, Rovšnik U, Hanke A, Martel A, Howard RJ, Lindahl E
|
| RgGuinier |
5.2 |
nm |
| Dmax |
16.7 |
nm |
| VolumePorod |
540 |
nm3 |
|
|
UniProt ID: O15530 (2-556) 3-phosphoinositide-dependent protein kinase 1
|
|
|

|
| Sample: |
3-phosphoinositide-dependent protein kinase 1 monomer, 65 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl pH 7.4, 250 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 26
|
Modulation of the substrate specificity of the kinase PDK1 by distinct conformations of the full-length protein
Science Signaling 16(789) (2023)
Sacerdoti M, Gross L, Riley A, Zehnder K, Ghode A, Klinke S, Anand G, Paris K, Winkel A, Herbrand A, Godage H, Cozier G, Süß E, Schulze J, Pastor-Flores D, Bollini M, Cappellari M, Svergun D, Gräwert M, Aramendia P, Leroux A, Potter B, Camacho C, Biondi R
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
107 |
nm3 |
|
|
UniProt ID: O15530 (2-556) 3-phosphoinositide-dependent protein kinase 1
UniProt ID: None (None-None) 2-O-benzoyl-Ins(1,3,4,5,6)P5
|
|
|

|
| Sample: |
3-phosphoinositide-dependent protein kinase 1 monomer, 65 kDa Homo sapiens protein
2-O-benzoyl-Ins(1,3,4,5,6)P5 monomer, 1 kDa synthetic construct
|
| Buffer: |
20 mM Tris-HCl pH 7.4, 250 mM NaCl, 1 mM DTT, 1 μM HYG8, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 26
|
Modulation of the substrate specificity of the kinase PDK1 by distinct conformations of the full-length protein
Science Signaling 16(789) (2023)
Sacerdoti M, Gross L, Riley A, Zehnder K, Ghode A, Klinke S, Anand G, Paris K, Winkel A, Herbrand A, Godage H, Cozier G, Süß E, Schulze J, Pastor-Flores D, Bollini M, Cappellari M, Svergun D, Gräwert M, Aramendia P, Leroux A, Potter B, Camacho C, Biondi R
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
98 |
nm3 |
|
|
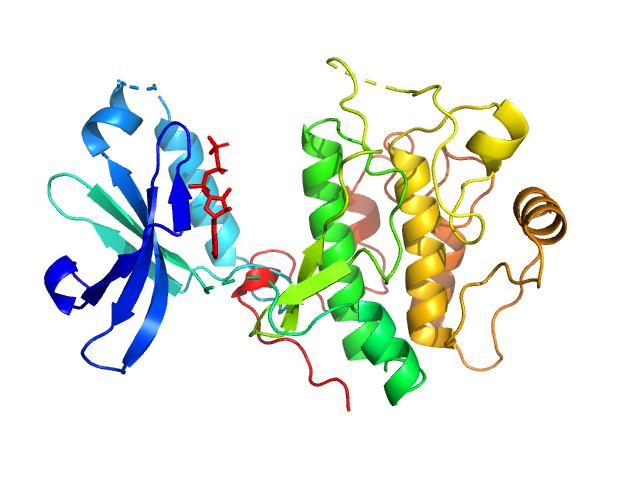
UniProt ID: O15530 (50-359) 3-phosphoinositide-dependent protein kinase 1 (Y188G Q292A)
|
|
|
|
| Sample: |
3-phosphoinositide-dependent protein kinase 1 (Y188G Q292A) monomer, 35 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl pH 7.4, 250 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 26
|
Modulation of the substrate specificity of the kinase PDK1 by distinct conformations of the full-length protein
Science Signaling 16(789) (2023)
Sacerdoti M, Gross L, Riley A, Zehnder K, Ghode A, Klinke S, Anand G, Paris K, Winkel A, Herbrand A, Godage H, Cozier G, Süß E, Schulze J, Pastor-Flores D, Bollini M, Cappellari M, Svergun D, Gräwert M, Aramendia P, Leroux A, Potter B, Camacho C, Biondi R
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
57 |
nm3 |
|
|
UniProt ID: Q9HXY5 (23-168) Skp-like protein
|
|
|
|
| Sample: |
Skp-like protein trimer, 55 kDa Pseudomonas aeruginosa (strain … protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 10 % glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 Q-Xoom, Center for Structural Studies, Heinrich-Heine-University on 2021 Feb 26
|
The periplasmic chaperone Skp prevents misfolding of the secretory lipase A from Pseudomonas aeruginosa
Frontiers in Molecular Biosciences 9 (2022)
Papadopoulos A, Busch M, Reiners J, Hachani E, Baeumers M, Berger J, Schmitt L, Jaeger K, Kovacic F, Smits S, Kedrov A
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
172 |
nm3 |
|
|
UniProt ID: Q9BXU0 (49-123) Testis-expressed protein 12
|
|
|

|
| Sample: |
Testis-expressed protein 12 dimer, 18 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jan 10
|
Coiled-coil structure of meiosis protein TEX12 and conformational regulation by its C-terminal tip
Communications Biology 5(1) (2022)
Dunce J, Salmon L, Davies O
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.6 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
UniProt ID: Q9BXU0 (49-113) Testis-expressed protein 12
|
|
|

|
| Sample: |
Testis-expressed protein 12 tetramer, 32 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jan 10
|
Coiled-coil structure of meiosis protein TEX12 and conformational regulation by its C-terminal tip
Communications Biology 5(1) (2022)
Dunce J, Salmon L, Davies O
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
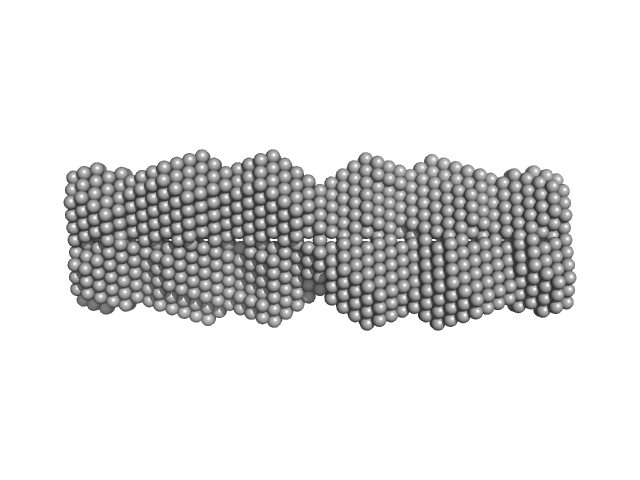
UniProt ID: Q9BXU0 (49-123) Testis-expressed protein 12 (L110E, F114E, I117E, L121E)
|
|
|
|
| Sample: |
Testis-expressed protein 12 (L110E, F114E, I117E, L121E) tetramer, 36 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Sep 15
|
Coiled-coil structure of meiosis protein TEX12 and conformational regulation by its C-terminal tip
Communications Biology 5(1) (2022)
Dunce J, Salmon L, Davies O
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.9 |
nm |
| VolumePorod |
59 |
nm3 |
|
|