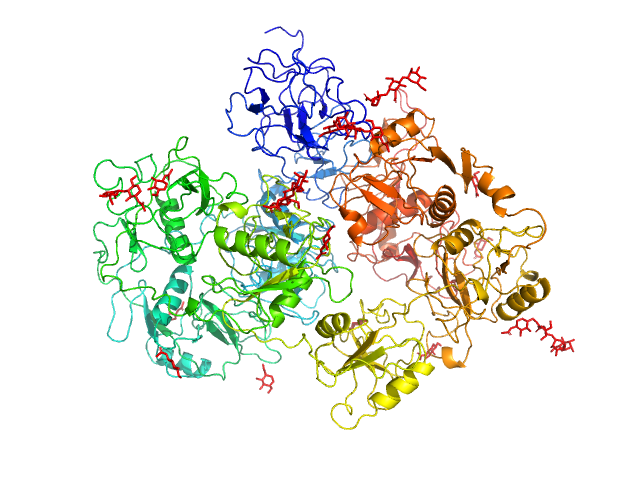
UniProt ID: Q13018 (21-1397) Secretory phospholipase A2 receptor
|
|
|
|
| Sample: |
Secretory phospholipase A2 receptor monomer, 189 kDa Homo sapiens protein
|
| Buffer: |
10 mM Bis-Tris, 150 mM NaCl, pH: 6.2 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 19
|
Structure of PLA2R reveals presentation of the dominant membranous nephropathy epitope and an immunogenic patch
Proceedings of the National Academy of Sciences 119(29) (2022)
Fresquet M, Lockhart-Cairns M, Rhoden S, Jowitt T, Briggs D, Baldock C, Brenchley P, Lennon R
|
| RgGuinier |
4.2 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
426 |
nm3 |
|
|
UniProt ID: P35555 (1-722) Fibrillin-1 PF3
|
|
|

|
| Sample: |
Fibrillin-1 PF3 monomer, 78 kDa Homo sapiens protein
|
| Buffer: |
Tris buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Dec 12
|
Latent TGFβ complexes are transglutaminase cross-linked to fibrillin to facilitate TGFβ activation.
Matrix Biol (2022)
Lockhart-Cairns MP, Cain SA, Dajani R, Steer R, Thomson J, Alanazi YF, Kielty CM, Baldock C
|
| RgGuinier |
5.4 |
nm |
| Dmax |
24.0 |
nm |
| VolumePorod |
141 |
nm3 |
|
|
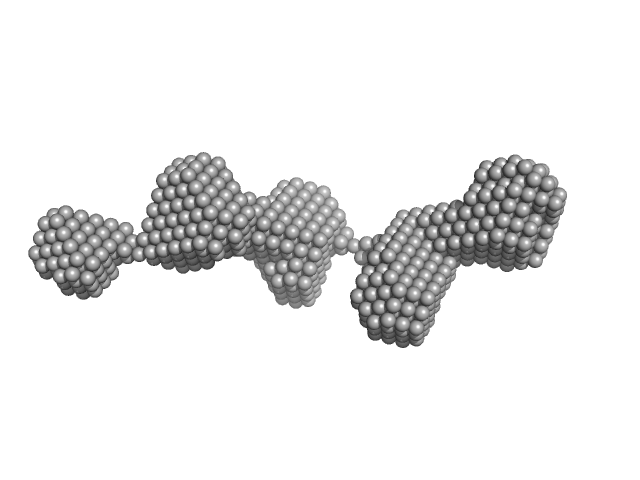
UniProt ID: Q14766-2 (1329-1721) Latent-transforming growth factor beta-binding protein 1
|
|
|
|
| Sample: |
Latent-transforming growth factor beta-binding protein 1 monomer, 44 kDa Homo sapiens protein
|
| Buffer: |
Tris buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Dec 12
|
Latent TGFβ complexes are transglutaminase cross-linked to fibrillin to facilitate TGFβ activation.
Matrix Biol (2022)
Lockhart-Cairns MP, Cain SA, Dajani R, Steer R, Thomson J, Alanazi YF, Kielty CM, Baldock C
|
| RgGuinier |
4.7 |
nm |
| Dmax |
20.5 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
UniProt ID: Q14766-2 (1329-1721) Latent-transforming growth factor beta-binding protein 1
UniProt ID: P35555 (1-722) Fibrillin-1 PF3
|
|
|

|
| Sample: |
Latent-transforming growth factor beta-binding protein 1 monomer, 44 kDa Homo sapiens protein
Fibrillin-1 PF3 monomer, 78 kDa Homo sapiens protein
|
| Buffer: |
10 mM Hepes, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Jul 20
|
Latent TGFβ complexes are transglutaminase cross-linked to fibrillin to facilitate TGFβ activation.
Matrix Biol (2022)
Lockhart-Cairns MP, Cain SA, Dajani R, Steer R, Thomson J, Alanazi YF, Kielty CM, Baldock C
|
| RgGuinier |
7.4 |
nm |
| Dmax |
29.6 |
nm |
| VolumePorod |
759 |
nm3 |
|
|
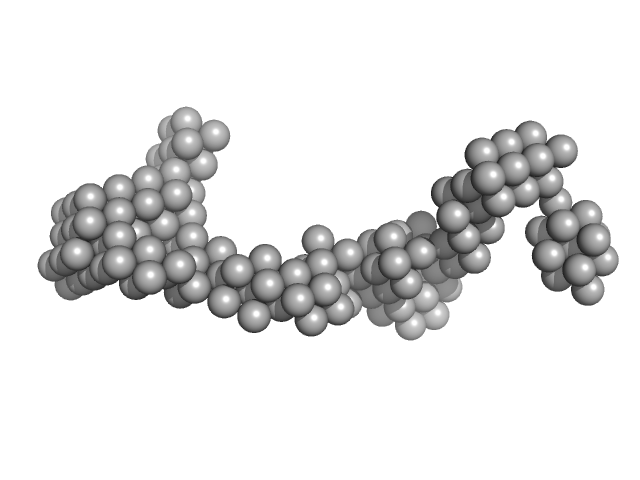
UniProt ID: P35555 (330-722) Fibrillin-1
|
|
|
|
| Sample: |
Fibrillin-1 monomer, 44 kDa Homo sapiens protein
|
| Buffer: |
Tris buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at ID02, ESRF on 2006 Feb 11
|
Transglutaminase-Mediated Cross-Linking of Tropoelastin to Fibrillin Stabilises the Elastin Precursor Prior to Elastic Fibre Assembly.
J Mol Biol 432(21):5736-5751 (2020)
Lockhart-Cairns MP, Newandee H, Thomson J, Weiss AS, Baldock C, Tarakanova A
|
| RgGuinier |
4.3 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
UniProt ID: P15502-2 (27-724) Elastin
|
|
|
|
| Sample: |
Elastin monomer, 60 kDa Homo sapiens protein
|
| Buffer: |
Phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at ID02, ESRF on 2011 Mar 15
|
Transglutaminase-Mediated Cross-Linking of Tropoelastin to Fibrillin Stabilises the Elastin Precursor Prior to Elastic Fibre Assembly.
J Mol Biol 432(21):5736-5751 (2020)
Lockhart-Cairns MP, Newandee H, Thomson J, Weiss AS, Baldock C, Tarakanova A
|
| RgGuinier |
6.6 |
nm |
| Dmax |
22.5 |
nm |
|
|
UniProt ID: P35555 (330-722) Fibrillin-1
UniProt ID: P15502 (27-724) Elastin
|
|
|
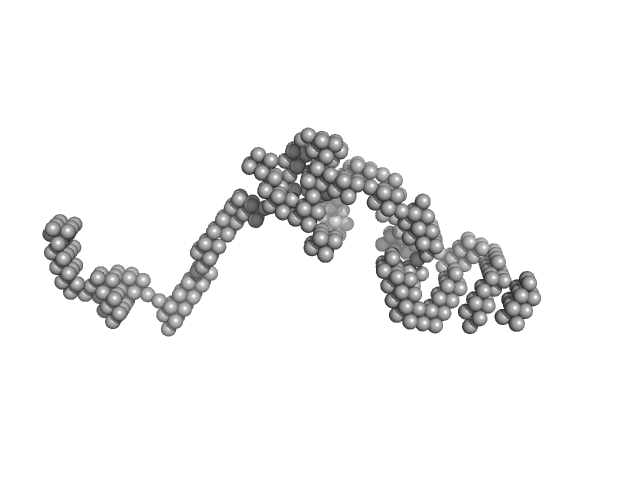
|
| Sample: |
Fibrillin-1 monomer, 44 kDa Homo sapiens protein
Elastin monomer, 60 kDa Homo sapiens protein
|
| Buffer: |
Tris buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Feb 21
|
Transglutaminase-Mediated Cross-Linking of Tropoelastin to Fibrillin Stabilises the Elastin Precursor Prior to Elastic Fibre Assembly.
J Mol Biol 432(21):5736-5751 (2020)
Lockhart-Cairns MP, Newandee H, Thomson J, Weiss AS, Baldock C, Tarakanova A
|
| RgGuinier |
7.1 |
nm |
| Dmax |
23.5 |
nm |
|
|
UniProt ID: P0DP23 (2-149) Calmodulin-1
|
|
|
|
| Sample: |
Calmodulin-1 monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM Hepes, 150 mM NaCl, 2 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Nov 15
|
Dynamics and structural changes of calmodulin upon interaction with the antagonist calmidazolium.
BMC Biol 20(1):176 (2022)
Léger C, Pitard I, Sadi M, Carvalho N, Brier S, Mechaly A, Raoux-Barbot D, Davi M, Hoos S, Weber P, Vachette P, Durand D, Haouz A, Guijarro JI, Ladant D, Chenal A
|
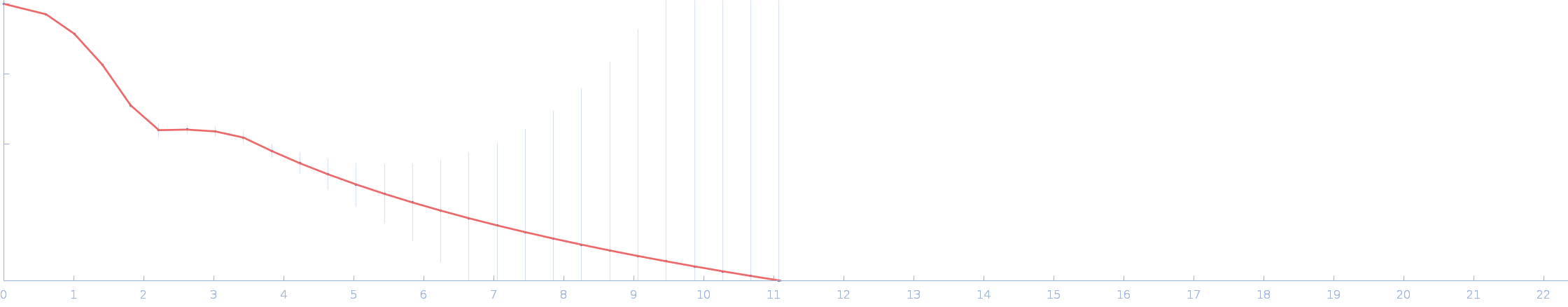
| RgGuinier |
2.2 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
UniProt ID: P0DP23 (2-149) Calmodulin-1
UniProt ID: None (None-None) Calmidazolium
|
|
|
|
| Sample: |
Calmodulin-1 monomer, 17 kDa Homo sapiens protein
Calmidazolium monomer, 1 kDa
|
| Buffer: |
20 mM Hepes, 150 mM NaCl, 2 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Nov 15
|
Dynamics and structural changes of calmodulin upon interaction with the antagonist calmidazolium.
BMC Biol 20(1):176 (2022)
Léger C, Pitard I, Sadi M, Carvalho N, Brier S, Mechaly A, Raoux-Barbot D, Davi M, Hoos S, Weber P, Vachette P, Durand D, Haouz A, Guijarro JI, Ladant D, Chenal A
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.2 |
nm |
| VolumePorod |
30 |
nm3 |
|
|
UniProt ID: O75469 (130-434) Nuclear receptor subfamily 1 group I member 2
UniProt ID: Q14994 (103-352) Nuclear receptor subfamily 1 group I member 3
|
|
|
|
| Sample: |
Nuclear receptor subfamily 1 group I member 2 monomer, 39 kDa Homo sapiens protein
Nuclear receptor subfamily 1 group I member 3 monomer, 32 kDa Homo sapiens protein
|
| Buffer: |
25 mM Hepes, 150 mM NaCl, 5% glycerol, 5 mM DTT, pH: 7.9 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2021 Oct 8
|
Molecular basis of crosstalk in nuclear receptors: heterodimerization between PXR and CAR and the implication in gene regulation
Shirish chodankar
|
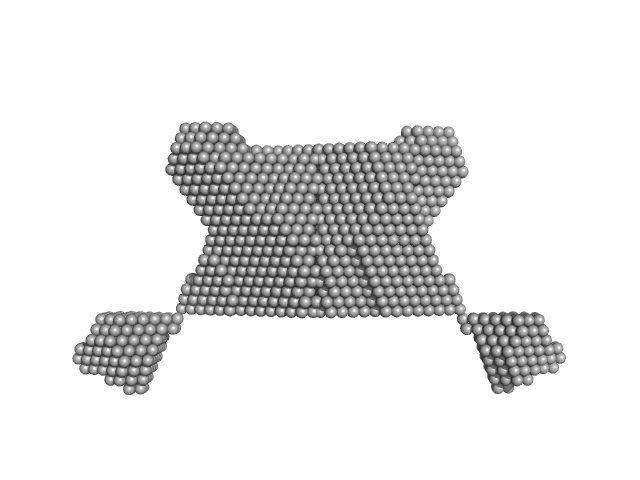
| RgGuinier |
2.7 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
56 |
nm3 |
|
|