UniProt ID: K9N4V7 (1-411) Nucleoprotein
UniProt ID: None (None-None) 5-(Propoxy)-1H-indole
|
|
|
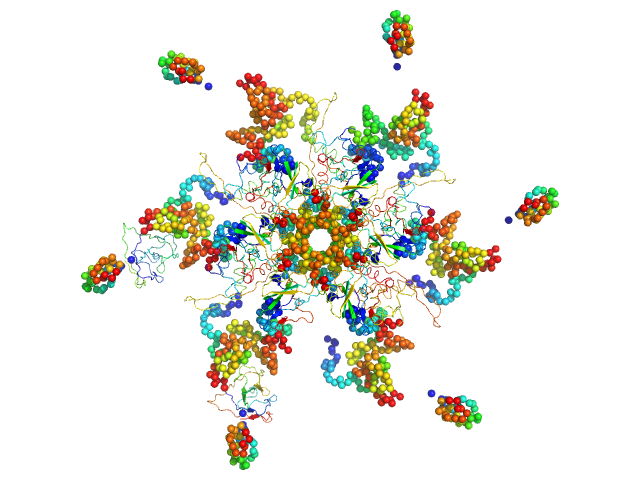
|
| Sample: |
Nucleoprotein dodecamer, 548 kDa Middle East respiratory … protein
5-(Propoxy)-1H-indole dodecamer, 2 kDa
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, pH: 8.5 |
| Experiment: |
SAXS
data collected at 23A, Taiwan Photon Source, NSRRC on 2019 Nov 22
|
Targeting the N-Terminus Domain of the Coronavirus Nucleocapsid Protein Induces Abnormal Oligomerization via Allosteric Modulation
Frontiers in Molecular Biosciences 9 (2022)
Hsu J, Chen J, Lin S, Hong J, Chen Y, Jeng U, Luo S, Hou M
|
| RgGuinier |
6.4 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
896 |
nm3 |
|
|
UniProt ID: K9N4V7 (1-411) Nucleoprotein
UniProt ID: None (None-None) 5-Isopropoxy-1H-indole
|
|
|
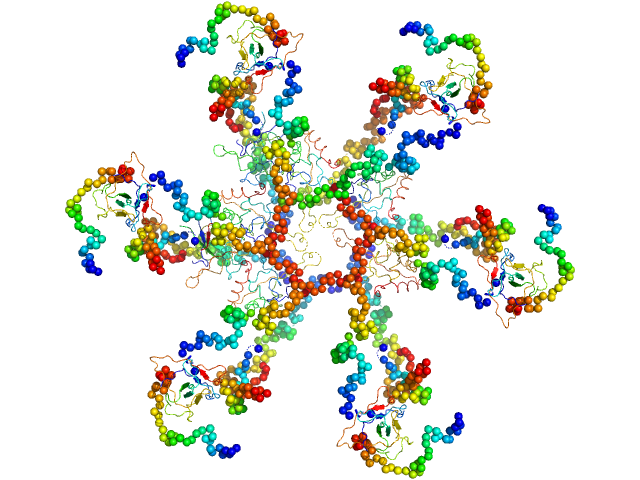
|
| Sample: |
Nucleoprotein dodecamer, 548 kDa Middle East respiratory … protein
5-Isopropoxy-1H-indole dodecamer, 2 kDa
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, pH: 8.5 |
| Experiment: |
SAXS
data collected at 23A, Taiwan Photon Source, NSRRC on 2019 Nov 22
|
Targeting the N-Terminus Domain of the Coronavirus Nucleocapsid Protein Induces Abnormal Oligomerization via Allosteric Modulation
Frontiers in Molecular Biosciences 9 (2022)
Hsu J, Chen J, Lin S, Hong J, Chen Y, Jeng U, Luo S, Hou M
|
| RgGuinier |
6.4 |
nm |
| Dmax |
22.2 |
nm |
| VolumePorod |
911 |
nm3 |
|
|
UniProt ID: K9N4V7 (1-411) Nucleoprotein
UniProt ID: None (None-None) 5-(2-fluoroethoxy)-1H-indole
|
|
|
|
| Sample: |
Nucleoprotein tetramer, 183 kDa Middle East respiratory … protein
5-(2-fluoroethoxy)-1H-indole tetramer, 1 kDa
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, pH: 8.5 |
| Experiment: |
SAXS
data collected at 23A, Taiwan Photon Source, NSRRC on 2019 Nov 26
|
Targeting the N-Terminus Domain of the Coronavirus Nucleocapsid Protein Induces Abnormal Oligomerization via Allosteric Modulation
Frontiers in Molecular Biosciences 9 (2022)
Hsu J, Chen J, Lin S, Hong J, Chen Y, Jeng U, Luo S, Hou M
|
| RgGuinier |
5.7 |
nm |
| Dmax |
19.1 |
nm |
| VolumePorod |
477 |
nm3 |
|
|
UniProt ID: K9N4V7 (1-411) Nucleoprotein
UniProt ID: None (None-None) 5-(2-methoxyethoxy)-1H-indole
|
|
|
|
| Sample: |
Nucleoprotein tetramer, 183 kDa Middle East respiratory … protein
5-(2-methoxyethoxy)-1H-indole tetramer, 1 kDa
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, pH: 8.5 |
| Experiment: |
SAXS
data collected at 23A, Taiwan Photon Source, NSRRC on 2019 Nov 26
|
Targeting the N-Terminus Domain of the Coronavirus Nucleocapsid Protein Induces Abnormal Oligomerization via Allosteric Modulation
Frontiers in Molecular Biosciences 9 (2022)
Hsu J, Chen J, Lin S, Hong J, Chen Y, Jeng U, Luo S, Hou M
|
| RgGuinier |
6.0 |
nm |
| Dmax |
18.5 |
nm |
| VolumePorod |
500 |
nm3 |
|
|
UniProt ID: None (None-None) Retinoblastoma-associated protein
|
|
|
|
| Sample: |
Retinoblastoma-associated protein monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate pH 7.0, 200 mM NaCl, 1mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 14
|
Conformational buffering underlies functional selection in intrinsically disordered protein regions.
Nat Struct Mol Biol (2022)
González-Foutel NS, Glavina J, Borcherds WM, Safranchik M, Barrera-Vilarmau S, Sagar A, Estaña A, Barozet A, Garrone NA, Fernandez-Ballester G, Blanes-Mira C, Sánchez IE, de Prat-Gay G, Cortés J, Bernadó P, Pappu RV, Holehouse AS, Daughdrill GW, Chemes LB
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
UniProt ID: None (None-None) Retinoblastoma-associated protein
|
|
|
|
| Sample: |
Retinoblastoma-associated protein monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate pH 7.0, 200 mM NaCl, 1mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 14
|
Conformational buffering underlies functional selection in intrinsically disordered protein regions.
Nat Struct Mol Biol (2022)
González-Foutel NS, Glavina J, Borcherds WM, Safranchik M, Barrera-Vilarmau S, Sagar A, Estaña A, Barozet A, Garrone NA, Fernandez-Ballester G, Blanes-Mira C, Sánchez IE, de Prat-Gay G, Cortés J, Bernadó P, Pappu RV, Holehouse AS, Daughdrill GW, Chemes LB
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.9 |
nm |
| VolumePorod |
64 |
nm3 |
|
|
UniProt ID: None (None-None) Retinoblastoma-associated protein
|
|
|
|
| Sample: |
Retinoblastoma-associated protein monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate pH 7.0, 200 mM NaCl, 1mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 14
|
Conformational buffering underlies functional selection in intrinsically disordered protein regions.
Nat Struct Mol Biol (2022)
González-Foutel NS, Glavina J, Borcherds WM, Safranchik M, Barrera-Vilarmau S, Sagar A, Estaña A, Barozet A, Garrone NA, Fernandez-Ballester G, Blanes-Mira C, Sánchez IE, de Prat-Gay G, Cortés J, Bernadó P, Pappu RV, Holehouse AS, Daughdrill GW, Chemes LB
|
| RgGuinier |
2.6 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
UniProt ID: None (None-None) Early E1A protein
|
|
|
|
| Sample: |
Early E1A protein monomer, 13 kDa Human adenovirus C … protein
|
| Buffer: |
20 mM sodium phosphate pH 7.0, 200 mM NaCl, 1mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 24
|
Conformational buffering underlies functional selection in intrinsically disordered protein regions.
Nat Struct Mol Biol (2022)
González-Foutel NS, Glavina J, Borcherds WM, Safranchik M, Barrera-Vilarmau S, Sagar A, Estaña A, Barozet A, Garrone NA, Fernandez-Ballester G, Blanes-Mira C, Sánchez IE, de Prat-Gay G, Cortés J, Bernadó P, Pappu RV, Holehouse AS, Daughdrill GW, Chemes LB
|
|
|
UniProt ID: None (None-None) Early E1A protein
|
|
|
|
| Sample: |
Early E1A protein monomer, 13 kDa Human adenovirus C … protein
|
| Buffer: |
20 mM sodium phosphate pH 7.0, 200 mM NaCl, 1mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 24
|
Conformational buffering underlies functional selection in intrinsically disordered protein regions.
Nat Struct Mol Biol (2022)
González-Foutel NS, Glavina J, Borcherds WM, Safranchik M, Barrera-Vilarmau S, Sagar A, Estaña A, Barozet A, Garrone NA, Fernandez-Ballester G, Blanes-Mira C, Sánchez IE, de Prat-Gay G, Cortés J, Bernadó P, Pappu RV, Holehouse AS, Daughdrill GW, Chemes LB
|
|
|
UniProt ID: None (None-None) Early E1A protein
|
|
|
|
| Sample: |
Early E1A protein monomer, 13 kDa Human adenovirus C … protein
|
| Buffer: |
20 mM sodium phosphate pH 7.0, 200 mM NaCl, 1mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 24
|
Conformational buffering underlies functional selection in intrinsically disordered protein regions.
Nat Struct Mol Biol (2022)
González-Foutel NS, Glavina J, Borcherds WM, Safranchik M, Barrera-Vilarmau S, Sagar A, Estaña A, Barozet A, Garrone NA, Fernandez-Ballester G, Blanes-Mira C, Sánchez IE, de Prat-Gay G, Cortés J, Bernadó P, Pappu RV, Holehouse AS, Daughdrill GW, Chemes LB
|
|
|