UniProt ID: None (None-None) Retinoblastoma-associated protein
UniProt ID: None (None-None) Early E1A protein
|
|
|
|
| Sample: |
Retinoblastoma-associated protein monomer, 41 kDa Homo sapiens protein
Early E1A protein monomer, 13 kDa Human adenovirus C … protein
|
| Buffer: |
20 mM sodium phosphate pH 7.0, 200 mM NaCl, 1mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 14
|
Conformational buffering underlies functional selection in intrinsically disordered protein regions.
Nat Struct Mol Biol (2022)
González-Foutel NS, Glavina J, Borcherds WM, Safranchik M, Barrera-Vilarmau S, Sagar A, Estaña A, Barozet A, Garrone NA, Fernandez-Ballester G, Blanes-Mira C, Sánchez IE, de Prat-Gay G, Cortés J, Bernadó P, Pappu RV, Holehouse AS, Daughdrill GW, Chemes LB
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
80 |
nm3 |
|
|
UniProt ID: None (None-None) Retinoblastoma-associated protein
UniProt ID: None (None-None) Early E1A protein
|
|
|
|
| Sample: |
Retinoblastoma-associated protein monomer, 41 kDa Homo sapiens protein
Early E1A protein monomer, 13 kDa Human adenovirus C … protein
|
| Buffer: |
20 mM sodium phosphate pH 7.0, 200 mM NaCl, 1mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 14
|
Conformational buffering underlies functional selection in intrinsically disordered protein regions.
Nat Struct Mol Biol (2022)
González-Foutel NS, Glavina J, Borcherds WM, Safranchik M, Barrera-Vilarmau S, Sagar A, Estaña A, Barozet A, Garrone NA, Fernandez-Ballester G, Blanes-Mira C, Sánchez IE, de Prat-Gay G, Cortés J, Bernadó P, Pappu RV, Holehouse AS, Daughdrill GW, Chemes LB
|
| RgGuinier |
3.0 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
UniProt ID: None (None-None) Retinoblastoma-associated protein
UniProt ID: None (None-None) Early E1A protein
|
|
|
|
| Sample: |
Retinoblastoma-associated protein monomer, 41 kDa Homo sapiens protein
Early E1A protein monomer, 13 kDa Human adenovirus C … protein
|
| Buffer: |
20 mM sodium phosphate pH 7.0, 200 mM NaCl, 1mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 14
|
Conformational buffering underlies functional selection in intrinsically disordered protein regions.
Nat Struct Mol Biol (2022)
González-Foutel NS, Glavina J, Borcherds WM, Safranchik M, Barrera-Vilarmau S, Sagar A, Estaña A, Barozet A, Garrone NA, Fernandez-Ballester G, Blanes-Mira C, Sánchez IE, de Prat-Gay G, Cortés J, Bernadó P, Pappu RV, Holehouse AS, Daughdrill GW, Chemes LB
|
| RgGuinier |
3.3 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
93 |
nm3 |
|
|
UniProt ID: None (None-None) Retinoblastoma-associated protein
UniProt ID: None (None-None) Early E1A protein
|
|
|
|
| Sample: |
Retinoblastoma-associated protein monomer, 41 kDa Homo sapiens protein
Early E1A protein monomer, 13 kDa Human adenovirus C … protein
|
| Buffer: |
20 mM sodium phosphate pH 7.0, 200 mM NaCl, 1mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 14
|
Conformational buffering underlies functional selection in intrinsically disordered protein regions.
Nat Struct Mol Biol (2022)
González-Foutel NS, Glavina J, Borcherds WM, Safranchik M, Barrera-Vilarmau S, Sagar A, Estaña A, Barozet A, Garrone NA, Fernandez-Ballester G, Blanes-Mira C, Sánchez IE, de Prat-Gay G, Cortés J, Bernadó P, Pappu RV, Holehouse AS, Daughdrill GW, Chemes LB
|
| RgGuinier |
3.0 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
UniProt ID: None (None-None) Early E1A protein
|
|
|
|
| Sample: |
Early E1A protein monomer, 13 kDa Human adenovirus C … protein
|
| Buffer: |
20 mM sodium phosphate pH 7.0, 200 mM NaCl, 1mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 24
|
Conformational buffering underlies functional selection in intrinsically disordered protein regions.
Nat Struct Mol Biol (2022)
González-Foutel NS, Glavina J, Borcherds WM, Safranchik M, Barrera-Vilarmau S, Sagar A, Estaña A, Barozet A, Garrone NA, Fernandez-Ballester G, Blanes-Mira C, Sánchez IE, de Prat-Gay G, Cortés J, Bernadó P, Pappu RV, Holehouse AS, Daughdrill GW, Chemes LB
|
| RgGuinier |
3.6 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
45 |
nm3 |
|
|
UniProt ID: A9ZTI0 (17-512) Apoptosis inducing protein
|
|
|
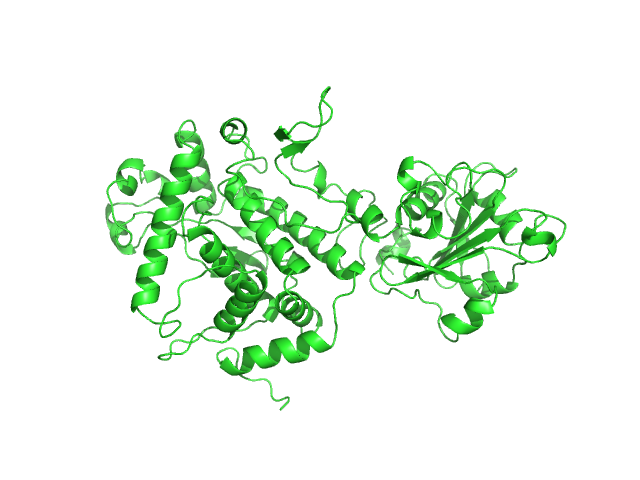
|
| Sample: |
Apoptosis inducing protein monomer, 57 kDa Photobacterium damselae subsp. … protein
|
| Buffer: |
50 mM Hepes, 500 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Apr 13
|
Unconventional structure and mechanisms for membrane interaction and translocation of the NF-κB-targeting toxin AIP56.
Nat Commun 14(1):7431 (2023)
Lisboa J, Pereira C, Pinto RD, Rodrigues IS, Pereira LMG, Pinheiro B, Oliveira P, Pereira PJB, Azevedo JE, Durand D, Benz R, do Vale A, Dos Santos NMS
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
UniProt ID: Q564N5 (23-594) Beta-galactosidase-like enzyme
|
|
|
|
| Sample: |
Beta-galactosidase-like enzyme dimer, 129 kDa Hamamotoa singularis protein
|
| Buffer: |
5 mM sodium phosphate, pH: 5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, Oak Ridge National Laboratory on 2018 Jun 25
|
Structural analysis and functional evaluation of the disordered ß–hexosyltransferase region from Hamamotoa (Sporobolomyces) singularis
Frontiers in Bioengineering and Biotechnology 11 (2023)
Dagher S, Vaishnav A, Stanley C, Meilleur F, Edwards B, Bruno-Bárcena J
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.4 |
nm |
| VolumePorod |
238 |
nm3 |
|
|
UniProt ID: Q564N5 (23-594) Beta-galactosidase-like enzyme
|
|
|
|
| Sample: |
Beta-galactosidase-like enzyme dimer, 129 kDa Hamamotoa singularis protein
|
| Buffer: |
5 mM sodium phosphate, pH: 5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, Oak Ridge National Laboratory on 2018 Jun 25
|
Structural analysis and functional evaluation of the disordered ß–hexosyltransferase region from Hamamotoa (Sporobolomyces) singularis
Frontiers in Bioengineering and Biotechnology 11 (2023)
Dagher S, Vaishnav A, Stanley C, Meilleur F, Edwards B, Bruno-Bárcena J
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
236 |
nm3 |
|
|
UniProt ID: Q01105 (1-225) SET nuclear proto-oncogene
|
|
|
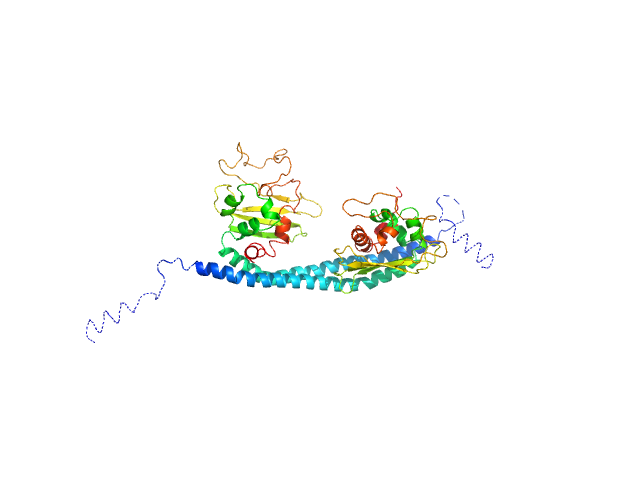
|
| Sample: |
SET nuclear proto-oncogene dimer, 53 kDa Homo sapiens protein
|
| Buffer: |
Sodium phosphate buffer, pH: 6.3 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 11
|
PP2A is activated by cytochrome c upon formation of a diffuse encounter complex with SET/TAF-Iβ
Computational and Structural Biotechnology Journal (2022)
Casado-Combreras M, Rivero-Rodríguez F, Elena-Real C, Molodenskiy D, Díaz-Quintana A, Martinho M, Gerbaud G, González-Arzola K, Velázquez-Campoy A, Svergun D, Belle V, De la Rosa M, Díaz-Moreno I
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
UniProt ID: P99999 (None-None) Cytochrome c
UniProt ID: Q01105 (1-225) SET nuclear proto-oncogene
|
|
|
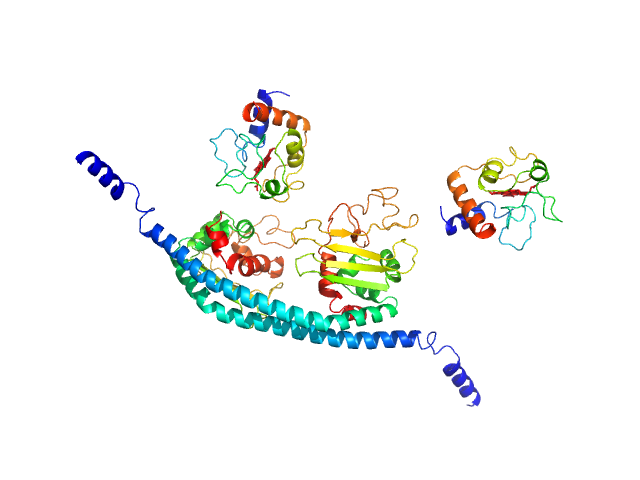
|
| Sample: |
Cytochrome c dimer, 24 kDa Homo sapiens protein
SET nuclear proto-oncogene dimer, 53 kDa Homo sapiens protein
|
| Buffer: |
Sodium phosphate buffer, pH: 6.3 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 11
|
PP2A is activated by cytochrome c upon formation of a diffuse encounter complex with SET/TAF-Iβ
Computational and Structural Biotechnology Journal (2022)
Casado-Combreras M, Rivero-Rodríguez F, Elena-Real C, Molodenskiy D, Díaz-Quintana A, Martinho M, Gerbaud G, González-Arzola K, Velázquez-Campoy A, Svergun D, Belle V, De la Rosa M, Díaz-Moreno I
|
| RgGuinier |
4.2 |
nm |
| Dmax |
16.0 |
nm |
|
|