UniProt ID: P01266 (1-2768) Thyroglobulin
|
|
|

|
| Sample: |
Thyroglobulin dimer, 610 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline, 150mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2020 Jul 17
|
Thyroglobulin - human and bovine SAXS/WAXS data
James Byrnes
|
| RgGuinier |
7.7 |
nm |
| Dmax |
26.3 |
nm |
| VolumePorod |
1800 |
nm3 |
|
|
UniProt ID: P01267 (1-2769) Thyroglobulin
|
|
|

|
| Sample: |
Thyroglobulin dimer, 606 kDa Bos taurus protein
|
| Buffer: |
phosphate buffered saline, 150mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2020 Jul 17
|
Thyroglobulin - human and bovine SAXS/WAXS data
James Byrnes
|
| RgGuinier |
8.1 |
nm |
| Dmax |
29.8 |
nm |
| VolumePorod |
1950 |
nm3 |
|
|
UniProt ID: P10205 (275-535) Tegument protein UL21 (C-terminal domain)
|
|
|

|
| Sample: |
Tegument protein UL21 (C-terminal domain) monomer, 29 kDa Human herpesvirus 1 … protein
|
| Buffer: |
20 mM Tris pH 8.5, 500 mM NaCl, 3% (v/v) glycerol, 1 mM DTT, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Sep 25
|
Herpes simplex virus 1 protein pUL21 alters ceramide metabolism by activating the inter-organelle transport protein CERT
Journal of Biological Chemistry :102589 (2022)
Benedyk T, Connor V, Caroe E, Shamin M, Svergun D, Deane J, Jeffries C, Crump C, Graham S
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
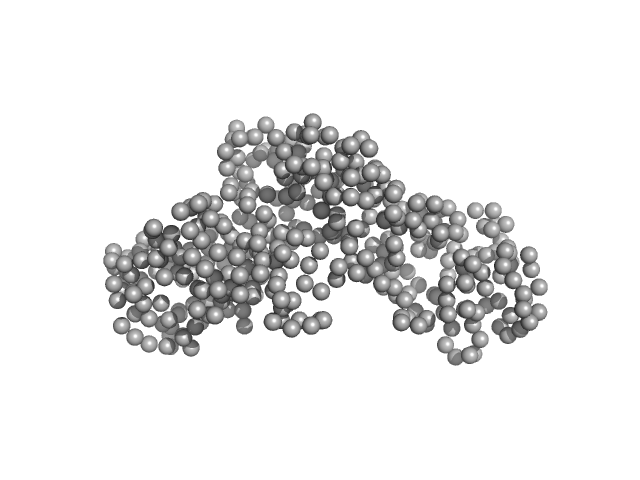
UniProt ID: Q9Y5P4 (None-None) Ceramide transfer protein (recombinant CERTL: amino acids 20-130 and 351-624)
|
|
|
|
| Sample: |
Ceramide transfer protein (recombinant CERTL: amino acids 20-130 and 351-624) monomer, 45 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8.5, 500 mM NaCl, 3% (v/v) glycerol, 1 mM DTT, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Sep 25
|
Herpes simplex virus 1 protein pUL21 alters ceramide metabolism by activating the inter-organelle transport protein CERT
Journal of Biological Chemistry :102589 (2022)
Benedyk T, Connor V, Caroe E, Shamin M, Svergun D, Deane J, Jeffries C, Crump C, Graham S
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
71 |
nm3 |
|
|
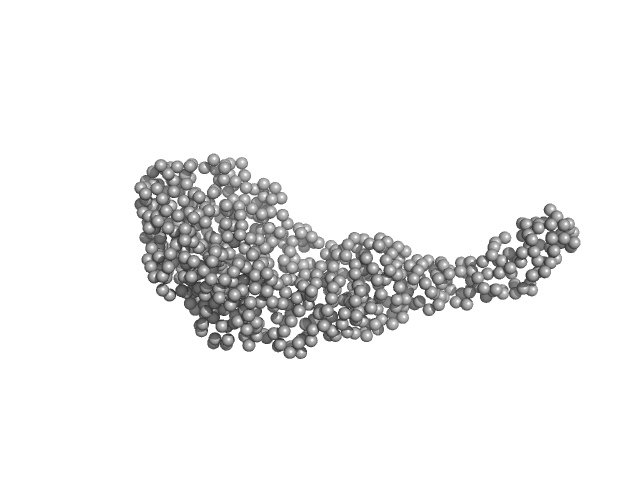
UniProt ID: P10205 (275-535) Tegument protein UL21 (C-terminal domain)
UniProt ID: Q9Y5P4 (None-None) Ceramide transfer protein (recombinant CERTL: amino acids 20-130 and 351-624)
|
|
|
|
| Sample: |
Tegument protein UL21 (C-terminal domain) monomer, 29 kDa Human herpesvirus 1 … protein
Ceramide transfer protein (recombinant CERTL: amino acids 20-130 and 351-624) monomer, 45 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8.5, 500 mM NaCl, 3% (v/v) glycerol, 1 mM DTT, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Sep 25
|
Herpes simplex virus 1 protein pUL21 alters ceramide metabolism by activating the inter-organelle transport protein CERT
Journal of Biological Chemistry :102589 (2022)
Benedyk T, Connor V, Caroe E, Shamin M, Svergun D, Deane J, Jeffries C, Crump C, Graham S
|
| RgGuinier |
3.3 |
nm |
| Dmax |
13.6 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
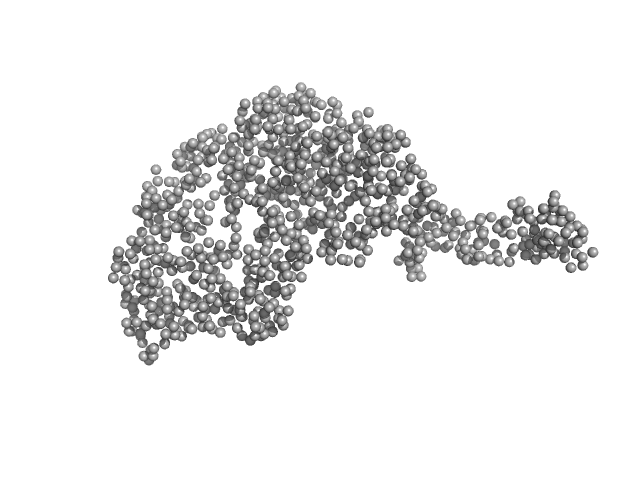
UniProt ID: P10205 (1-535) Tegument protein UL21
UniProt ID: Q9Y5P4 (None-None) Ceramide transfer protein (recombinant CERTL: amino acids 20-130 and 351-624)
|
|
|
|
| Sample: |
Tegument protein UL21 monomer, 59 kDa Human alphaherpesvirus 1 … protein
Ceramide transfer protein (recombinant CERTL: amino acids 20-130 and 351-624) monomer, 45 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8.5, 500 mM NaCl, 3% (v/v) glycerol, 1 mM DTT, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Sep 25
|
Herpes simplex virus 1 protein pUL21 alters ceramide metabolism by activating the inter-organelle transport protein CERT
Journal of Biological Chemistry :102589 (2022)
Benedyk T, Connor V, Caroe E, Shamin M, Svergun D, Deane J, Jeffries C, Crump C, Graham S
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
158 |
nm3 |
|
|
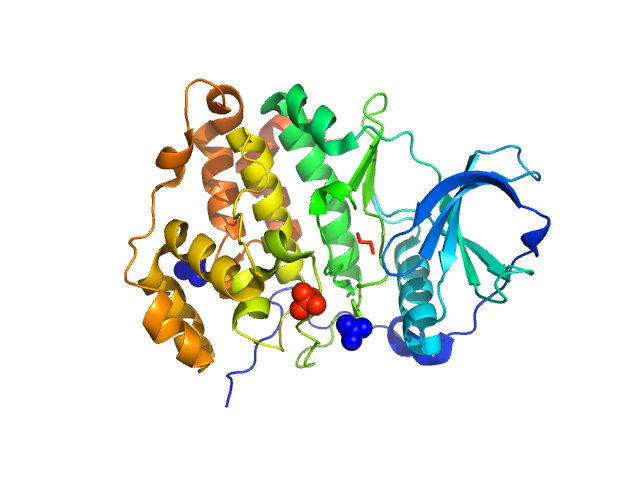
UniProt ID: P68400 (3-330) Casein kinase II subunit alpha
|
|
|
|
| Sample: |
Casein kinase II subunit alpha monomer, 39 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 500 mM NaCl, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Oct 10
|
Mechanism of CK2 Inhibition by a Ruthenium-Based Polyoxometalate.
Front Mol Biosci 9:906390 (2022)
Fabbian S, Giachin G, Bellanda M, Borgo C, Ruzzene M, Spuri G, Campofelice A, Veneziano L, Bonchio M, Carraro M, Battistutta R
|
| RgGuinier |
2.2 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
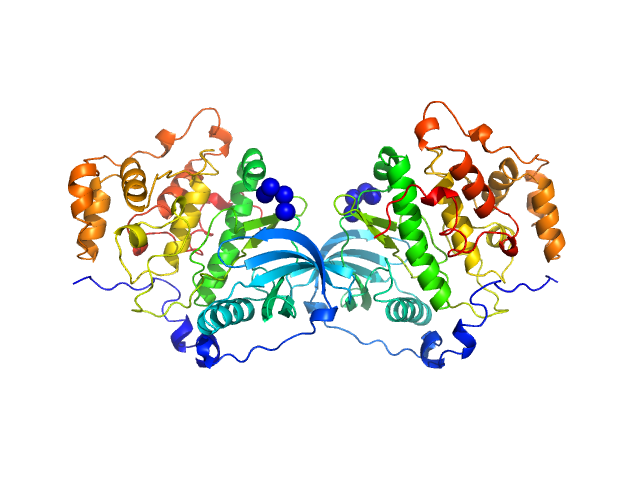
UniProt ID: P68400 (3-330) Casein kinase II subunit alpha
|
|
|
|
| Sample: |
Casein kinase II subunit alpha dimer, 78 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 500 mM NaCl, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Oct 10
|
Mechanism of CK2 Inhibition by a Ruthenium-Based Polyoxometalate.
Front Mol Biosci 9:906390 (2022)
Fabbian S, Giachin G, Bellanda M, Borgo C, Ruzzene M, Spuri G, Campofelice A, Veneziano L, Bonchio M, Carraro M, Battistutta R
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
122 |
nm3 |
|
|
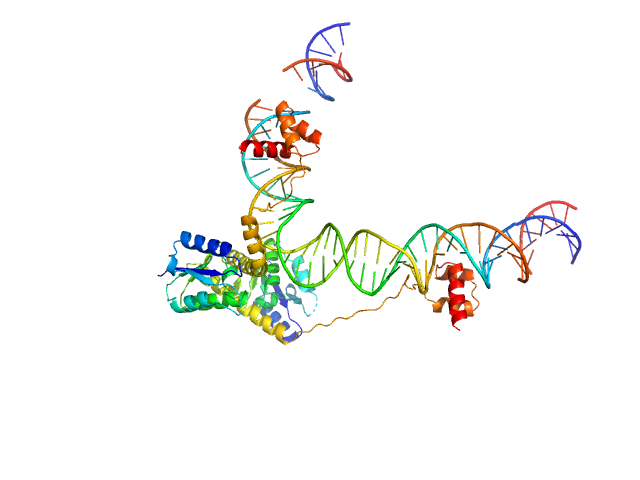
UniProt ID: P0ADI2 (1-185) Transposon Tn3 resolvase
UniProt ID: None (None-None) Tn3 res, resolvase binding site II
|
|
|
|
| Sample: |
Transposon Tn3 resolvase dimer, 43 kDa Escherichia coli protein
Tn3 res, resolvase binding site II monomer, 32 kDa DNA
|
| Buffer: |
25 mM Tris, 150 mM NaCl, 10 mM MgCl2, 1% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2014 Nov 6
|
Structural basis for topological regulation of Tn3 resolvase.
Nucleic Acids Res (2022)
Montaño SP, Rowland SJ, Fuller JR, Burke ME, MacDonald AI, Boocock MR, Stark WM, Rice PA
|
| RgGuinier |
5.0 |
nm |
| Dmax |
14.9 |
nm |
| VolumePorod |
217 |
nm3 |
|
|
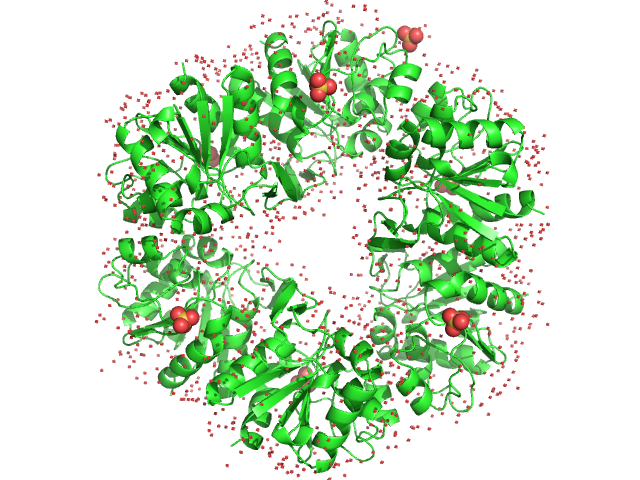
UniProt ID: O59413 (1-166) Deglycase PH1704
|
|
|
|
| Sample: |
Deglycase PH1704 hexamer, 112 kDa Pyrococcus horikoshii (strain … protein
|
| Buffer: |
20 mM Tris pH 7.5, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Jun 7
|
Medical contrast agents as promising tools for biomacromolecular SAXS experiments.
Acta Crystallogr D Struct Biol 78(Pt 9):1120-1130 (2022)
Gabel F, Engilberge S, Schmitt E, Thureau A, Mechulam Y, Pérez J, Girard E
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
140 |
nm3 |
|
|