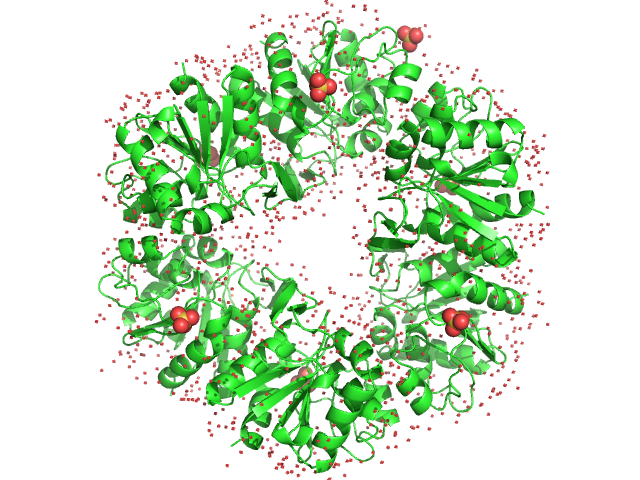
UniProt ID: O59413 (1-166) Deglycase PH1704
|
|
|
|
| Sample: |
Deglycase PH1704 hexamer, 112 kDa Pyrococcus horikoshii (strain … protein
|
| Buffer: |
20 mM Tris pH 7.5 and 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Nov 26
|
Medical contrast agents as promising tools for biomacromolecular SAXS experiments.
Acta Crystallogr D Struct Biol 78(Pt 9):1120-1130 (2022)
Gabel F, Engilberge S, Schmitt E, Thureau A, Mechulam Y, Pérez J, Girard E
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
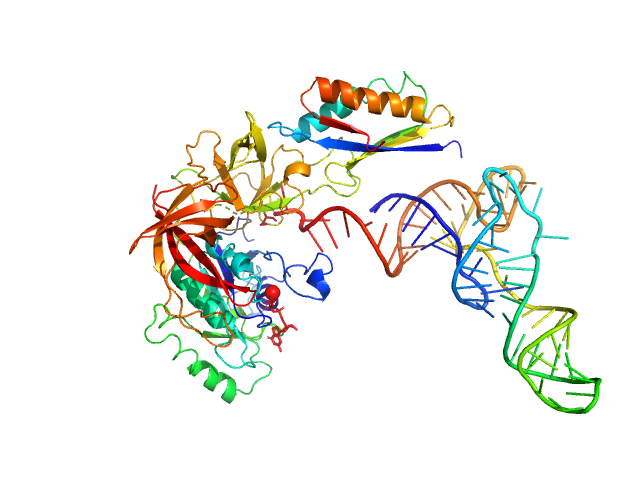
UniProt ID: Q980A5 (2-415) Translation initiation factor 2 subunit gamma
UniProt ID: Q97Z79 (None-None) Translation initiation factor 2 subunit alpha
UniProt ID: None (None-None) transfer RNA
|
|
|
|
| Sample: |
Translation initiation factor 2 subunit gamma monomer, 46 kDa Saccharolobus solfataricus (strain … protein
Translation initiation factor 2 subunit alpha monomer, 10 kDa Saccharolobus solfataricus (strain … protein
Transfer RNA monomer, 23 kDa Escherichia coli RNA
|
| Buffer: |
10 mM MOPS- NaOH pH 6.7, 200 mM NaCl, 5 mM MgCl 2, 1 mM GDPNP, pH: 6.7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Nov 26
|
Medical contrast agents as promising tools for biomacromolecular SAXS experiments.
Acta Crystallogr D Struct Biol 78(Pt 9):1120-1130 (2022)
Gabel F, Engilberge S, Schmitt E, Thureau A, Mechulam Y, Pérez J, Girard E
|
| RgGuinier |
3.6 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
UniProt ID: A0A100HLL5 (10-214) DNA protection during starvation, DPS (Ferritin superfamily)
|
|
|

|
| Sample: |
DNA protection during starvation, DPS (Ferritin superfamily) dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 50 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 22
|
The Conformation of the N-Terminal Tails of Deinococcus grandis Dps Is Modulated by the Ionic Strength
International Journal of Molecular Sciences 23(9):4871 (2022)
Guerra J, Blanchet C, Vieira B, Almeida A, Waerenborgh J, Jones N, Hoffmann S, Tavares P, Pereira A
|
| RgGuinier |
4.4 |
nm |
| Dmax |
16.1 |
nm |
| VolumePorod |
409 |
nm3 |
|
|
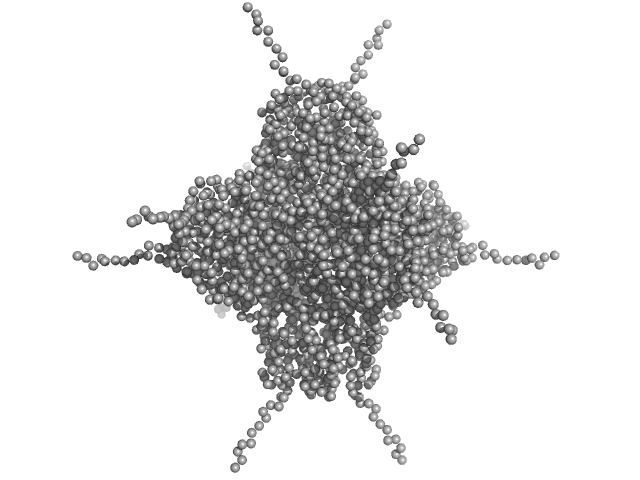
UniProt ID: A0A100HLL5 (10-214) DNA protection during starvation, DPS (Ferritin superfamily)
|
|
|
|
| Sample: |
DNA protection during starvation, DPS (Ferritin superfamily) dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 80 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 22
|
The Conformation of the N-Terminal Tails of Deinococcus grandis Dps Is Modulated by the Ionic Strength
International Journal of Molecular Sciences 23(9):4871 (2022)
Guerra J, Blanchet C, Vieira B, Almeida A, Waerenborgh J, Jones N, Hoffmann S, Tavares P, Pereira A
|
| RgGuinier |
4.5 |
nm |
| Dmax |
17.1 |
nm |
| VolumePorod |
475 |
nm3 |
|
|
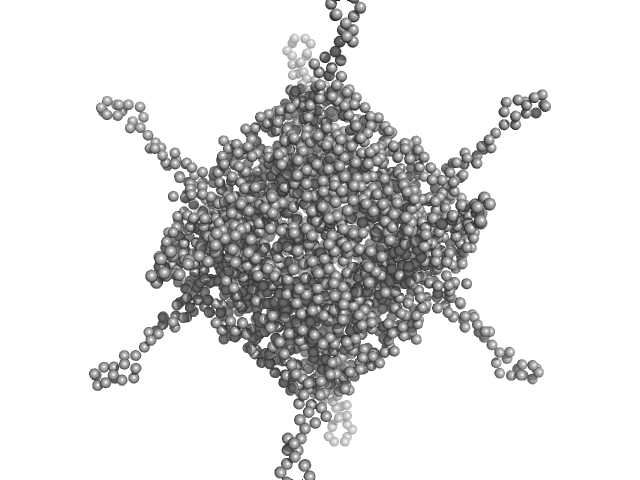
UniProt ID: A0A100HLL5 (10-214) DNA protection during starvation, DPS (Ferritin superfamily)
|
|
|
|
| Sample: |
DNA protection during starvation, DPS (Ferritin superfamily) dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 230 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 22
|
The Conformation of the N-Terminal Tails of Deinococcus grandis Dps Is Modulated by the Ionic Strength
International Journal of Molecular Sciences 23(9):4871 (2022)
Guerra J, Blanchet C, Vieira B, Almeida A, Waerenborgh J, Jones N, Hoffmann S, Tavares P, Pereira A
|
| RgGuinier |
4.5 |
nm |
| Dmax |
20.6 |
nm |
| VolumePorod |
438 |
nm3 |
|
|
UniProt ID: A0A100HLL5 (10-214) DNA protection during starvation, DPS (Ferritin superfamily)
|
|
|
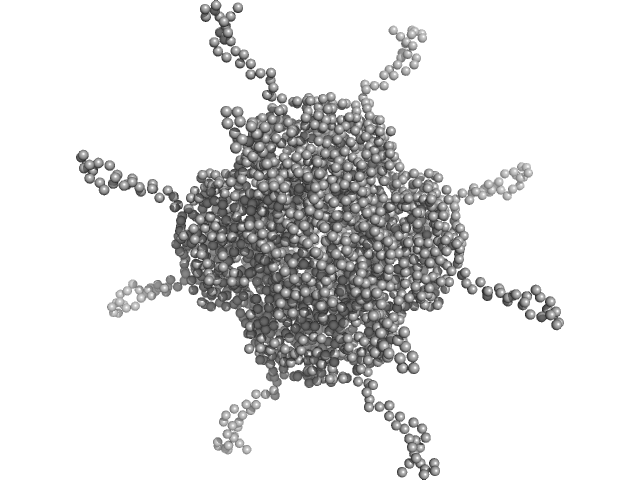
|
| Sample: |
DNA protection during starvation, DPS (Ferritin superfamily) dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 480 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 22
|
The Conformation of the N-Terminal Tails of Deinococcus grandis Dps Is Modulated by the Ionic Strength
International Journal of Molecular Sciences 23(9):4871 (2022)
Guerra J, Blanchet C, Vieira B, Almeida A, Waerenborgh J, Jones N, Hoffmann S, Tavares P, Pereira A
|
| RgGuinier |
4.8 |
nm |
| Dmax |
20.5 |
nm |
| VolumePorod |
430 |
nm3 |
|
|
UniProt ID: A0A100HLL5 (56-214) DNA protection during starvation, DPS-∆N (Ferritin superfamily)
|
|
|

|
| Sample: |
DNA protection during starvation, DPS-∆N (Ferritin superfamily) dodecamer, 218 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 50 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 22
|
The Conformation of the N-Terminal Tails of Deinococcus grandis Dps Is Modulated by the Ionic Strength
International Journal of Molecular Sciences 23(9):4871 (2022)
Guerra J, Blanchet C, Vieira B, Almeida A, Waerenborgh J, Jones N, Hoffmann S, Tavares P, Pereira A
|
| RgGuinier |
3.8 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
290 |
nm3 |
|
|
UniProt ID: A0A100HLL5 (56-214) DNA protection during starvation, DPS-∆N (Ferritin superfamily)
|
|
|

|
| Sample: |
DNA protection during starvation, DPS-∆N (Ferritin superfamily) dodecamer, 218 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 230 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 22
|
The Conformation of the N-Terminal Tails of Deinococcus grandis Dps Is Modulated by the Ionic Strength
International Journal of Molecular Sciences 23(9):4871 (2022)
Guerra J, Blanchet C, Vieira B, Almeida A, Waerenborgh J, Jones N, Hoffmann S, Tavares P, Pereira A
|
| RgGuinier |
3.8 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
291 |
nm3 |
|
|
UniProt ID: None (None-None) Glyco_trans_2-like domain-containing protein
|
|
|
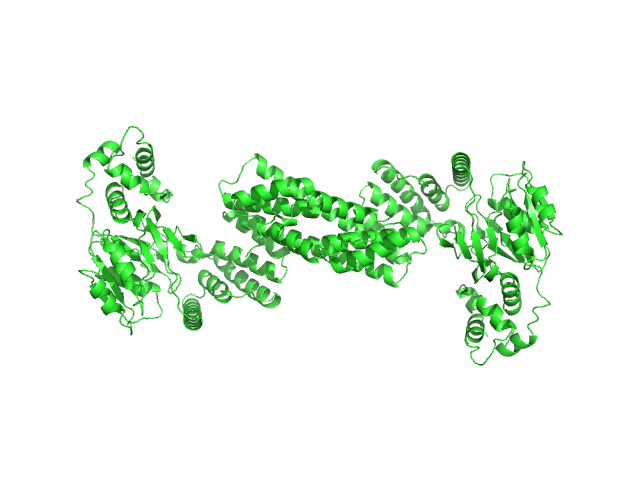
|
| Sample: |
Glyco_trans_2-like domain-containing protein dimer, 96 kDa Streptomyces sp. M41(2017) protein
|
| Buffer: |
20 mM Tris, pH 7.5, 100 mM NaCl, and 2 mM DTT + 1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2019 Oct 18
|
Global shape of SvGT, a metal-dependent bacteriocin modifying S/O-HexNActransferase from actinobacteria: c -terminal dimerization modulates the function of this GT.
J Biomol Struct Dyn :1-15 (2023)
Sharma Y, Ahlawat S, Ashish, Rao A
|
| RgGuinier |
4.4 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
151 |
nm3 |
|
|
UniProt ID: None (None-None) Glyco_trans_2-like domain-containing protein
|
|
|
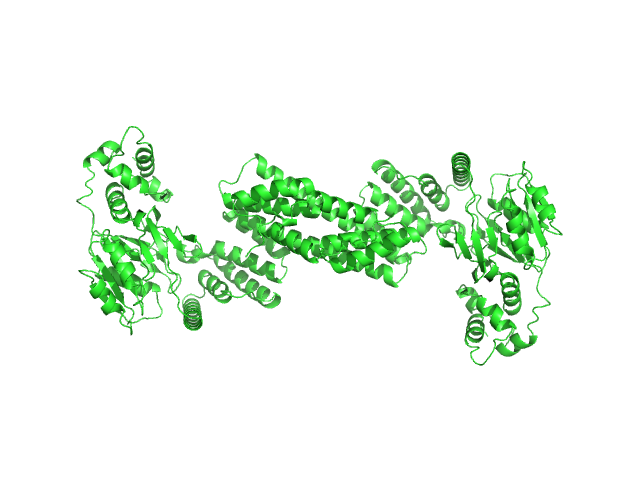
|
| Sample: |
Glyco_trans_2-like domain-containing protein dimer, 96 kDa Streptomyces sp. M41(2017) protein
|
| Buffer: |
20 mM Tris, pH 7.5, 100 mM NaCl, 2 mM DTT, 1 mM EDTA, 2 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2019 Oct 18
|
Global shape of SvGT, a metal-dependent bacteriocin modifying S/O-HexNActransferase from actinobacteria: c -terminal dimerization modulates the function of this GT.
J Biomol Struct Dyn :1-15 (2023)
Sharma Y, Ahlawat S, Ashish, Rao A
|
| RgGuinier |
5.8 |
nm |
| Dmax |
17.9 |
nm |
| VolumePorod |
111 |
nm3 |
|
|