UniProt ID: I6X8D2 (1154-1720) Polyketide synthase Pks13
|
|
|
|
| Sample: |
Polyketide synthase Pks13 monomer, 62 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris-HCl 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Nov 1
|
Solution structure of the type I polyketide synthase Pks13 from Mycobacterium tuberculosis.
BMC Biol 20(1):147 (2022)
Bon C, Cabantous S, Julien S, Guillet V, Chalut C, Rima J, Brison Y, Malaga W, Sanchez-Dafun A, Gavalda S, Quémard A, Marcoux J, Waldo GS, Guilhot C, Mourey L
|
| RgGuinier |
3.2 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
80 |
nm3 |
|
|
UniProt ID: I6X8D2 (576-1063) Polyketide synthase Pks13
|
|
|

|
| Sample: |
Polyketide synthase Pks13 monomer, 53 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris-HCl, 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Nov 1
|
Solution structure of the type I polyketide synthase Pks13 from Mycobacterium tuberculosis.
BMC Biol 20(1):147 (2022)
Bon C, Cabantous S, Julien S, Guillet V, Chalut C, Rima J, Brison Y, Malaga W, Sanchez-Dafun A, Gavalda S, Quémard A, Marcoux J, Waldo GS, Guilhot C, Mourey L
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
UniProt ID: I6X8D2 (591-1046) Polyketide synthase Pks13
|
|
|

|
| Sample: |
Polyketide synthase Pks13 monomer, 51 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris-HCl, 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2011 Jun 3
|
Solution structure of the type I polyketide synthase Pks13 from Mycobacterium tuberculosis.
BMC Biol 20(1):147 (2022)
Bon C, Cabantous S, Julien S, Guillet V, Chalut C, Rima J, Brison Y, Malaga W, Sanchez-Dafun A, Gavalda S, Quémard A, Marcoux J, Waldo GS, Guilhot C, Mourey L
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
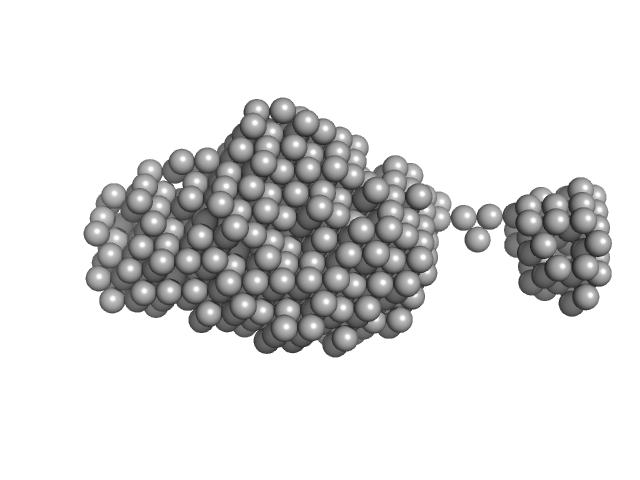
UniProt ID: I6X8D2 (1-1063) Polyketide synthase Pks13
|
|
|

|
| Sample: |
Polyketide synthase Pks13 monomer, 116 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris-HCl 50 mM NaCl, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Nov 1
|
Solution structure of the type I polyketide synthase Pks13 from Mycobacterium tuberculosis.
BMC Biol 20(1):147 (2022)
Bon C, Cabantous S, Julien S, Guillet V, Chalut C, Rima J, Brison Y, Malaga W, Sanchez-Dafun A, Gavalda S, Quémard A, Marcoux J, Waldo GS, Guilhot C, Mourey L
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.9 |
nm |
| VolumePorod |
186 |
nm3 |
|
|
UniProt ID: I6X8D2 (119-575) Polyketide synthase Pks13
|
|
|

|
| Sample: |
Polyketide synthase Pks13 monomer, 51 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris-HCl, 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2012 Dec 7
|
Solution structure of the type I polyketide synthase Pks13 from Mycobacterium tuberculosis.
BMC Biol 20(1):147 (2022)
Bon C, Cabantous S, Julien S, Guillet V, Chalut C, Rima J, Brison Y, Malaga W, Sanchez-Dafun A, Gavalda S, Quémard A, Marcoux J, Waldo GS, Guilhot C, Mourey L
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
82 |
nm3 |
|
|
UniProt ID: I6X8D2 (119-1070) Polyketide synthase Pks13
|
|
|

|
| Sample: |
Polyketide synthase Pks13 monomer, 104 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris-HCl, 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2008 Sep 20
|
Solution structure of the type I polyketide synthase Pks13 from Mycobacterium tuberculosis.
BMC Biol 20(1):147 (2022)
Bon C, Cabantous S, Julien S, Guillet V, Chalut C, Rima J, Brison Y, Malaga W, Sanchez-Dafun A, Gavalda S, Quémard A, Marcoux J, Waldo GS, Guilhot C, Mourey L
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
155 |
nm3 |
|
|
UniProt ID: Q02251 (1-2111) Mycocerosic acid synthase
|
|
|

|
| Sample: |
Mycocerosic acid synthase monomer, 224 kDa Mycobacterium bovis (strain … protein
|
| Buffer: |
50 mM Tris-HCl, 50 mM NaCl, 10% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2013 Oct 1
|
Solution structure of the type I polyketide synthase Pks13 from Mycobacterium tuberculosis.
BMC Biol 20(1):147 (2022)
Bon C, Cabantous S, Julien S, Guillet V, Chalut C, Rima J, Brison Y, Malaga W, Sanchez-Dafun A, Gavalda S, Quémard A, Marcoux J, Waldo GS, Guilhot C, Mourey L
|
| RgGuinier |
5.7 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
420 |
nm3 |
|
|
UniProt ID: A0A0H3M7U0 (1-1876) Phenolpthiocerol synthesis type-I polyketide synthase ppsA
|
|
|

|
| Sample: |
Phenolpthiocerol synthesis type-I polyketide synthase ppsA monomer, 199 kDa Mycobacterium bovis (strain … protein
|
| Buffer: |
50 mM Tris-HCl, 500 mM NaCl, 2 mM EDTA, 10% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2012 Jun 14
|
Solution structure of the type I polyketide synthase Pks13 from Mycobacterium tuberculosis.
BMC Biol 20(1):147 (2022)
Bon C, Cabantous S, Julien S, Guillet V, Chalut C, Rima J, Brison Y, Malaga W, Sanchez-Dafun A, Gavalda S, Quémard A, Marcoux J, Waldo GS, Guilhot C, Mourey L
|
| RgGuinier |
6.7 |
nm |
| Dmax |
20.2 |
nm |
| VolumePorod |
680 |
nm3 |
|
|
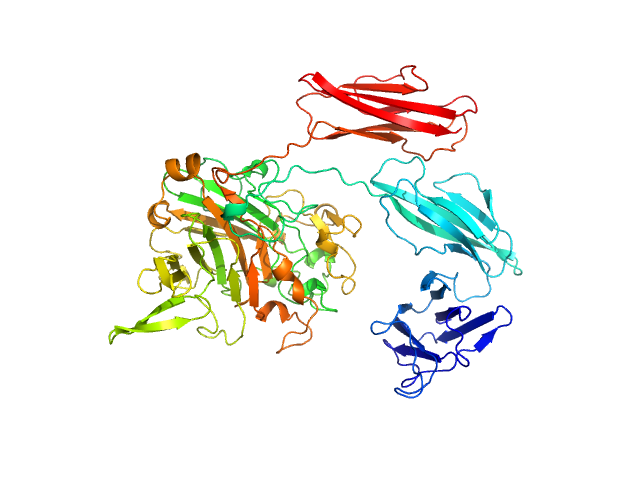
UniProt ID: A0A2N0UYM2 (31-665) Dockerin type I repeat
|
|
|
|
| Sample: |
Dockerin type I repeat monomer, 69 kDa Ruminococcus bromii protein
|
| Buffer: |
phosphate buffered saline, 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Nov 14
|
The Ruminococcus bromii amylosome protein Sas6 binds single and double helical α-glucan structures in starch.
Nat Struct Mol Biol (2024)
Photenhauer AL, Villafuerte-Vega RC, Cerqueira FM, Armbruster KM, Mareček F, Chen T, Wawrzak Z, Hopkins JB, Vander Kooi CW, Janeček Š, Ruotolo BT, Koropatkin NM
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
97 |
nm3 |
|
|
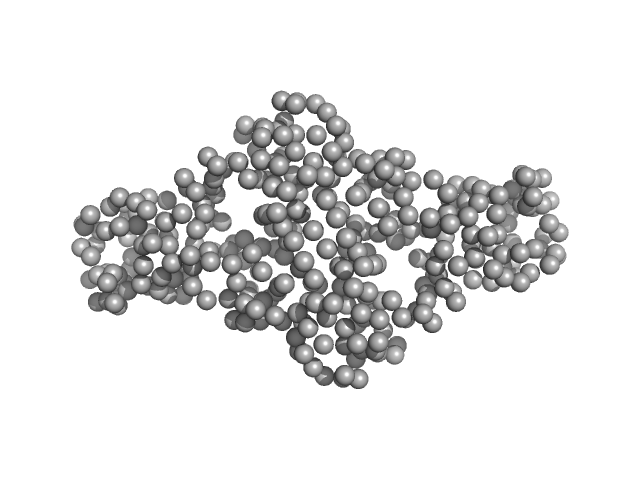
UniProt ID: Q07449 (44-201) Extracellular superoxide dismutase [Cu-Zn]
|
|
|
|
| Sample: |
Extracellular superoxide dismutase [Cu-Zn] dimer, 34 kDa Onchocerca volvulus protein
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, 2 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 13
|
Crystal structure of an extracellular superoxide dismutase from Onchocerca volvulus
and implications for parasite-specific drug development
Acta Crystallographica Section F Structural Biology Communications 78(6) (2022)
Moustafa A, Perbandt M, Liebau E, Betzel C, Falke S
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
55 |
nm3 |
|
|