UniProt ID: P32657 (1009-1274) chromodomain helicase DNA binding domain
|
|
|
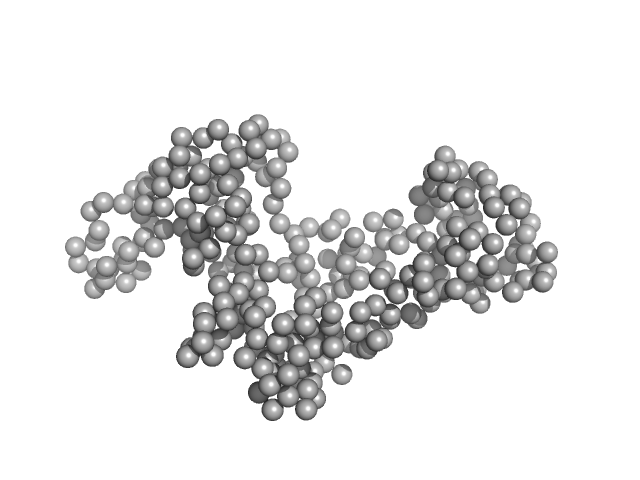
|
| Sample: |
Chromodomain helicase DNA binding domain monomer, 31 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
50mM Hepes 150mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Nov 20
|
Structural reorganization of the chromatin remodeling enzyme Chd1 upon engagement with nucleosomes.
Elife 6 (2017)
Sundaramoorthy R, Hughes AL, Singh V, Wiechens N, Ryan DP, El-Mkami H, Petoukhov M, Svergun DI, Treutlein B, Quack S, Fischer M, Michaelis J, Böttcher B, Norman DG, Owen-Hughes T
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.3 |
nm |
|
|
UniProt ID: P32657 (133-1305) chromodomain helicase DNA binding domain
|
|
|
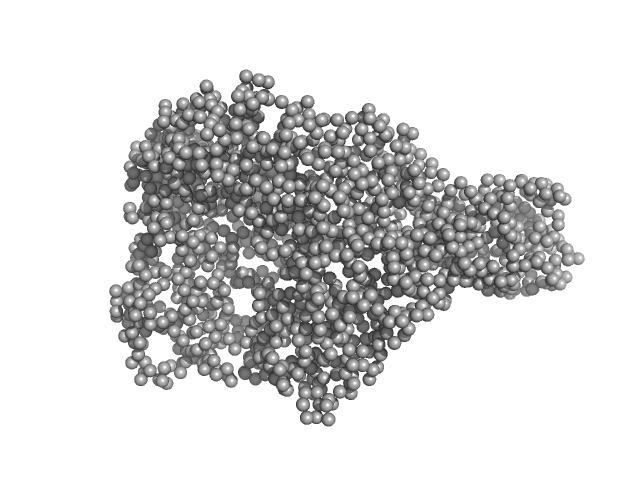
|
| Sample: |
Chromodomain helicase DNA binding domain monomer, 135 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
50mM Hepes 150mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2008 Nov 30
|
Structural reorganization of the chromatin remodeling enzyme Chd1 upon engagement with nucleosomes.
Elife 6 (2017)
Sundaramoorthy R, Hughes AL, Singh V, Wiechens N, Ryan DP, El-Mkami H, Petoukhov M, Svergun DI, Treutlein B, Quack S, Fischer M, Michaelis J, Böttcher B, Norman DG, Owen-Hughes T
|
| RgGuinier |
4.2 |
nm |
| Dmax |
15.4 |
nm |
| VolumePorod |
280 |
nm3 |
|
|
UniProt ID: P32657 (1-1305) chromodomain helicase DNA binding domain
|
|
|
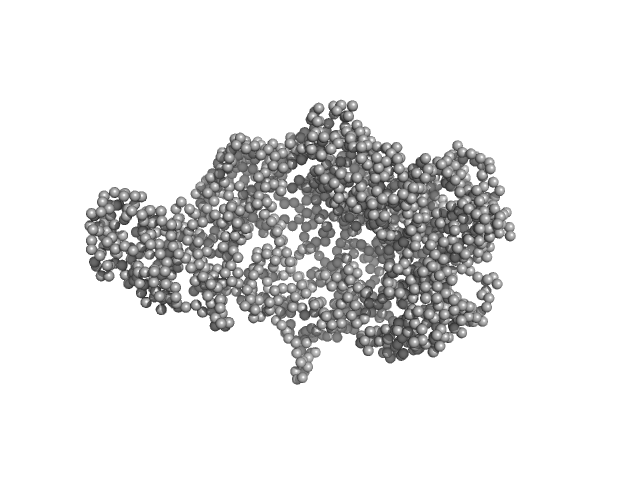
|
| Sample: |
Chromodomain helicase DNA binding domain monomer, 150 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
50mM Hepes 150mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2008 Nov 30
|
Structural reorganization of the chromatin remodeling enzyme Chd1 upon engagement with nucleosomes.
Elife 6 (2017)
Sundaramoorthy R, Hughes AL, Singh V, Wiechens N, Ryan DP, El-Mkami H, Petoukhov M, Svergun DI, Treutlein B, Quack S, Fischer M, Michaelis J, Böttcher B, Norman DG, Owen-Hughes T
|
| RgGuinier |
4.9 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
340 |
nm3 |
|
|
UniProt ID: P32657 (133-1010) chromodomain helicase DNA binding domain
|
|
|
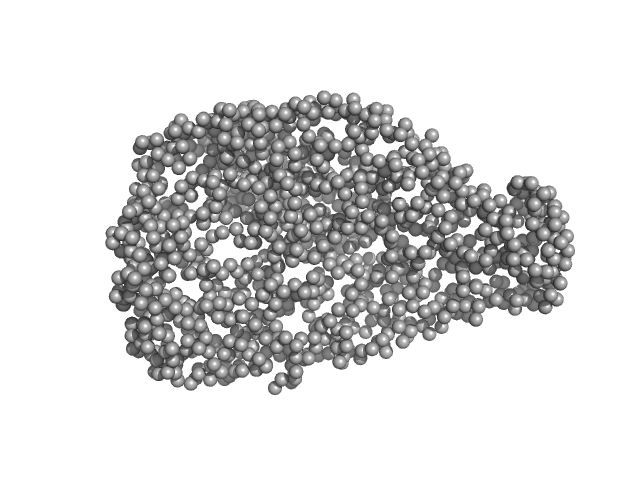
|
| Sample: |
Chromodomain helicase DNA binding domain monomer, 102 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
50mM Hepes 150mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2008 Nov 30
|
Structural reorganization of the chromatin remodeling enzyme Chd1 upon engagement with nucleosomes.
Elife 6 (2017)
Sundaramoorthy R, Hughes AL, Singh V, Wiechens N, Ryan DP, El-Mkami H, Petoukhov M, Svergun DI, Treutlein B, Quack S, Fischer M, Michaelis J, Böttcher B, Norman DG, Owen-Hughes T
|
| RgGuinier |
4.1 |
nm |
| Dmax |
16.1 |
nm |
| VolumePorod |
190 |
nm3 |
|
|
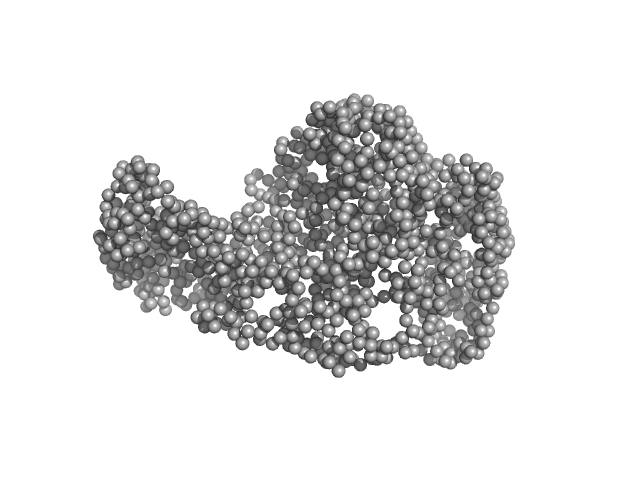
UniProt ID: P32657 (1-1010) chromodomain helicase DNA binding domain
|
|
|
|
| Sample: |
Chromodomain helicase DNA binding domain monomer, 117 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
50mM Hepes 150mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2008 Nov 30
|
Structural reorganization of the chromatin remodeling enzyme Chd1 upon engagement with nucleosomes.
Elife 6 (2017)
Sundaramoorthy R, Hughes AL, Singh V, Wiechens N, Ryan DP, El-Mkami H, Petoukhov M, Svergun DI, Treutlein B, Quack S, Fischer M, Michaelis J, Böttcher B, Norman DG, Owen-Hughes T
|
| RgGuinier |
4.5 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
228 |
nm3 |
|
|
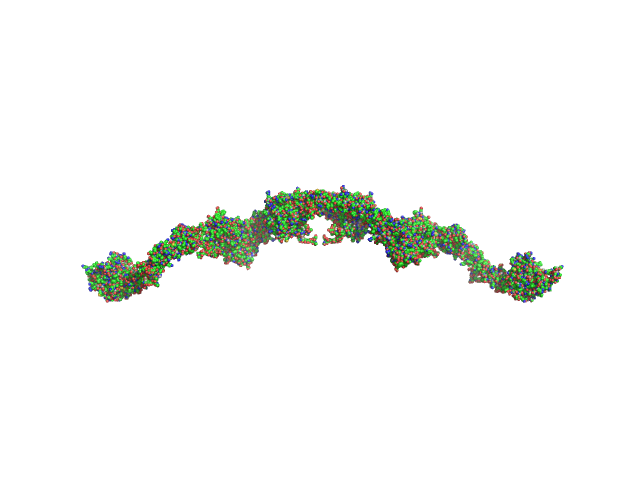
UniProt ID: P00736 (18-705) Complement C1r subcomponent
UniProt ID: P09871 (16-688) Complement C1s subcomponent
|
|
|
|
| Sample: |
Complement C1r subcomponent dimer, 156 kDa Homo sapiens protein
Complement C1s subcomponent dimer, 150 kDa Homo sapiens protein
|
| Buffer: |
50 mM TrisHCl, 145 mM NaCl, 3 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Dec 8
|
Structure and activation of C1, the complex initiating the classical pathway of the complement cascade.
Proc Natl Acad Sci U S A 114(5):986-991 (2017)
Mortensen SA, Sander B, Jensen RK, Pedersen JS, Golas MM, Jensenius JC, Hansen AG, Thiel S, Andersen GR
|
|
|
UniProt ID: P02747 (23-245) Complement C1q subcomponent subunit C
UniProt ID: P02746 (28-253) Complement C1q subcomponent subunit B
UniProt ID: P02745 (23-245) Complement C1q subcomponent subunit A
|
|
|
|
| Sample: |
Complement C1q subcomponent subunit C hexamer, 142 kDa Homo sapiens protein
Complement C1q subcomponent subunit B hexamer, 142 kDa Homo sapiens protein
Complement C1q subcomponent subunit A hexamer, 142 kDa Homo sapiens protein
|
| Buffer: |
50 mM TrisHCl, 145 mM NaCl, 3 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Dec 8
|
Structure and activation of C1, the complex initiating the classical pathway of the complement cascade.
Proc Natl Acad Sci U S A 114(5):986-991 (2017)
Mortensen SA, Sander B, Jensen RK, Pedersen JS, Golas MM, Jensenius JC, Hansen AG, Thiel S, Andersen GR
|
|
|
UniProt ID: P02747 (23-245) Complement C1q subcomponent subunit C
UniProt ID: P02746 (28-253) Complement C1q subcomponent subunit B
UniProt ID: P02745 (23-245) Complement C1q subcomponent subunit A
UniProt ID: P00736 (18-705) Complement C1r subcomponent
UniProt ID: P09871 (16-688) Complement C1s subcomponent
|
|
|

|
| Sample: |
Complement C1q subcomponent subunit C hexamer, 142 kDa Homo sapiens protein
Complement C1q subcomponent subunit B hexamer, 142 kDa Homo sapiens protein
Complement C1q subcomponent subunit A hexamer, 142 kDa Homo sapiens protein
Complement C1r subcomponent dimer, 156 kDa Homo sapiens protein
Complement C1s subcomponent dimer, 150 kDa Homo sapiens protein
|
| Buffer: |
50 mM EPPS, 145 mM NaCl, 3 mM CaCl2, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Aug 16
|
Structure and activation of C1, the complex initiating the classical pathway of the complement cascade.
Proc Natl Acad Sci U S A 114(5):986-991 (2017)
Mortensen SA, Sander B, Jensen RK, Pedersen JS, Golas MM, Jensenius JC, Hansen AG, Thiel S, Andersen GR
|
| RgGuinier |
11.5 |
nm |
| Dmax |
36.6 |
nm |
|
|
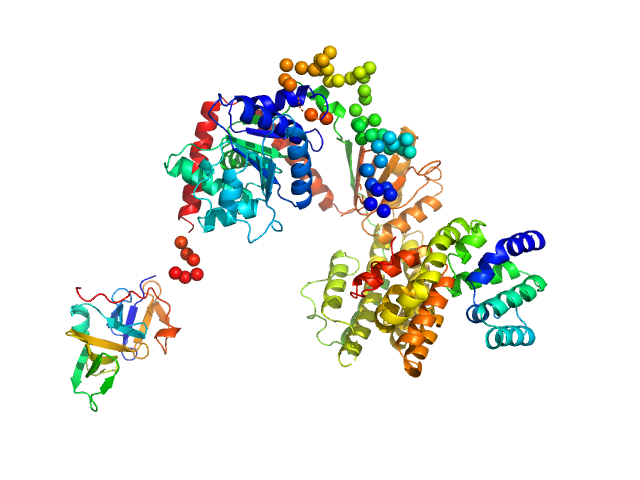
UniProt ID: O95786 (1-925) Probable ATP-dependent RNA helicase DDX58
|
|
|
|
| Sample: |
Probable ATP-dependent RNA helicase DDX58 monomer, 108 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2.5 mM MgCl2, 10% glycerol and 1mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2012 Apr 6
|
Combined roles of ATP and small hairpin RNA in the activation of RIG-I revealed by solution-based analysis.
Nucleic Acids Res 46(6):3169-3186 (2018)
Shah N, Beckham SA, Wilce JA, Wilce MCJ
|
| RgGuinier |
4.3 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
186 |
nm3 |
|
|
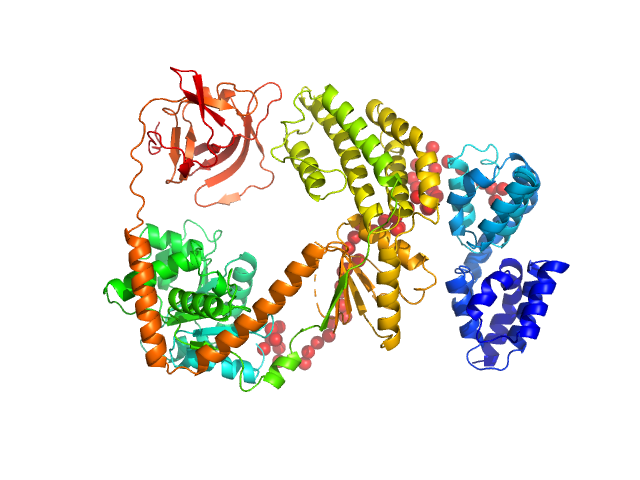
UniProt ID: O95786 (1-925) Probable ATP-dependent RNA helicase DDX58
|
|
|
|
| Sample: |
Probable ATP-dependent RNA helicase DDX58 monomer, 108 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2.5 mM MgCl2, 10% glycerol and 1mM DTT, 2mM ADP-AlFx, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Nov 20
|
Combined roles of ATP and small hairpin RNA in the activation of RIG-I revealed by solution-based analysis.
Nucleic Acids Res 46(6):3169-3186 (2018)
Shah N, Beckham SA, Wilce JA, Wilce MCJ
|
| RgGuinier |
4.2 |
nm |
| Dmax |
15.6 |
nm |
| VolumePorod |
190 |
nm3 |
|
|