UniProt ID: P0DTC1 (3943-4140) Replicase polyprotein 1a (Non-structural protein 8, SARS-CoV-2)
|
|
|
|
| Sample: |
Replicase polyprotein 1a (Non-structural protein 8, SARS-CoV-2) dimer, 44 kDa Severe acute respiratory … protein
|
| Buffer: |
20 mM HEPES pH 7.4, 250 mM NaCl, pH: 7.4 |
| Experiment: |
SANS
data collected at BL01-Small Angle Neutron Scattering, China Spallation Neutron Source on 2021 Jul 19
|
Multiscale characterization reveals oligomerization dependent phase separation of primer-independent RNA polymerase nsp8 from SARS-CoV-2.
Commun Biol 5(1):925 (2022)
Xu J, Jiang X, Zhang Y, Dong Y, Ma C, Jiang H, Zuo T, Chen R, Ke Y, Cheng H, Wang H, Liu J
|
|
|
UniProt ID: P0DTC1 (3943-4140) Replicase polyprotein 1a (Non-structural protein 8, SARS-CoV-2)
|
|
|
|
| Sample: |
Replicase polyprotein 1a (Non-structural protein 8, SARS-CoV-2) dimer, 44 kDa Severe acute respiratory … protein
|
| Buffer: |
20 mM HEPES pH 7.4, 500 mM NaCl, pH: 7.4 |
| Experiment: |
SANS
data collected at BL01-Small Angle Neutron Scattering, China Spallation Neutron Source on 2021 Jul 19
|
Multiscale characterization reveals oligomerization dependent phase separation of primer-independent RNA polymerase nsp8 from SARS-CoV-2.
Commun Biol 5(1):925 (2022)
Xu J, Jiang X, Zhang Y, Dong Y, Ma C, Jiang H, Zuo T, Chen R, Ke Y, Cheng H, Wang H, Liu J
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
98 |
nm3 |
|
|
UniProt ID: P0DTC1 (3943-4140) Replicase polyprotein 1a (Non-structural protein 8, SARS-CoV-2)
|
|
|
|
| Sample: |
Replicase polyprotein 1a (Non-structural protein 8, SARS-CoV-2) dimer, 44 kDa Severe acute respiratory … protein
|
| Buffer: |
20 mM HEPES pH 7.4, 1000 mM NaCl, pH: 7.4 |
| Experiment: |
SANS
data collected at BL01-Small Angle Neutron Scattering, China Spallation Neutron Source on 2021 Jul 19
|
Multiscale characterization reveals oligomerization dependent phase separation of primer-independent RNA polymerase nsp8 from SARS-CoV-2.
Commun Biol 5(1):925 (2022)
Xu J, Jiang X, Zhang Y, Dong Y, Ma C, Jiang H, Zuo T, Chen R, Ke Y, Cheng H, Wang H, Liu J
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
100 |
nm3 |
|
|
UniProt ID: P0DTC1 (None-None) Replicase polyprotein 1a (Non-structural protein 8, SARS-CoV-2)
|
|
|
|
| Sample: |
Replicase polyprotein 1a (Non-structural protein 8, SARS-CoV-2) tetramer, 88 kDa Severe acute respiratory … protein
|
| Buffer: |
20 mM HEPES pH 7.4, 250 mM NaCl, pH: 7.4 |
| Experiment: |
SANS
data collected at BL01-Small Angle Neutron Scattering, China Spallation Neutron Source on 2021 Jul 19
|
Multiscale characterization reveals oligomerization dependent phase separation of primer-independent RNA polymerase nsp8 from SARS-CoV-2.
Commun Biol 5(1):925 (2022)
Xu J, Jiang X, Zhang Y, Dong Y, Ma C, Jiang H, Zuo T, Chen R, Ke Y, Cheng H, Wang H, Liu J
|
| RgGuinier |
6.3 |
nm |
| Dmax |
24.0 |
nm |
|
|
UniProt ID: P0DTC1 (None-None) Replicase polyprotein 1a (Non-structural protein 8, SARS-CoV-2)
|
|
|
|
| Sample: |
Replicase polyprotein 1a (Non-structural protein 8, SARS-CoV-2) tetramer, 88 kDa Severe acute respiratory … protein
|
| Buffer: |
20 mM HEPES pH 7.4, 500 mM NaCl, pH: 7.4 |
| Experiment: |
SANS
data collected at BL01-Small Angle Neutron Scattering, China Spallation Neutron Source on 2021 Jul 19
|
Multiscale characterization reveals oligomerization dependent phase separation of primer-independent RNA polymerase nsp8 from SARS-CoV-2.
Commun Biol 5(1):925 (2022)
Xu J, Jiang X, Zhang Y, Dong Y, Ma C, Jiang H, Zuo T, Chen R, Ke Y, Cheng H, Wang H, Liu J
|
| RgGuinier |
5.2 |
nm |
| Dmax |
18.0 |
nm |
|
|
UniProt ID: P0DTC1 (None-None) Replicase polyprotein 1a (Non-structural protein 8, SARS-CoV-2)
|
|
|
|
| Sample: |
Replicase polyprotein 1a (Non-structural protein 8, SARS-CoV-2) tetramer, 88 kDa Severe acute respiratory … protein
|
| Buffer: |
20 mM HEPES pH 7.4, 1000 mM NaCl, pH: 7.4 |
| Experiment: |
SANS
data collected at BL01-Small Angle Neutron Scattering, China Spallation Neutron Source on 2021 Jul 19
|
Multiscale characterization reveals oligomerization dependent phase separation of primer-independent RNA polymerase nsp8 from SARS-CoV-2.
Commun Biol 5(1):925 (2022)
Xu J, Jiang X, Zhang Y, Dong Y, Ma C, Jiang H, Zuo T, Chen R, Ke Y, Cheng H, Wang H, Liu J
|
| RgGuinier |
4.8 |
nm |
| Dmax |
16.0 |
nm |
|
|
UniProt ID: P32004 (712-917) Neural cell adhesion molecule L1
|
|
|

|
| Sample: |
Neural cell adhesion molecule L1 monomer, 24 kDa Homo sapiens protein
|
| Buffer: |
50 mM MES pH 6.5, 150 mM NaCl, 5% glycerol, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 May 11
|
X‐ray structure and function of fibronectin domains two and three of the neural cell adhesion molecule L1
The FASEB Journal 37(3) (2023)
Guédez G, Loers G, Jeffries C, Kozak S, Meijers R, Svergun D, Schachner M, Löw C
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
UniProt ID: P32004 (712-917) Neural cell adhesion molecule L1
|
|
|

|
| Sample: |
Neural cell adhesion molecule L1 monomer, 24 kDa Homo sapiens protein
|
| Buffer: |
50 mM MES pH 6.5, 150 mM NaCl, 5% glycerol, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 May 11
|
X‐ray structure and function of fibronectin domains two and three of the neural cell adhesion molecule L1
The FASEB Journal 37(3) (2023)
Guédez G, Loers G, Jeffries C, Kozak S, Meijers R, Svergun D, Schachner M, Löw C
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
72 |
nm3 |
|
|
UniProt ID: P32004 (712-917) Neural cell adhesion molecule L1
|
|
|
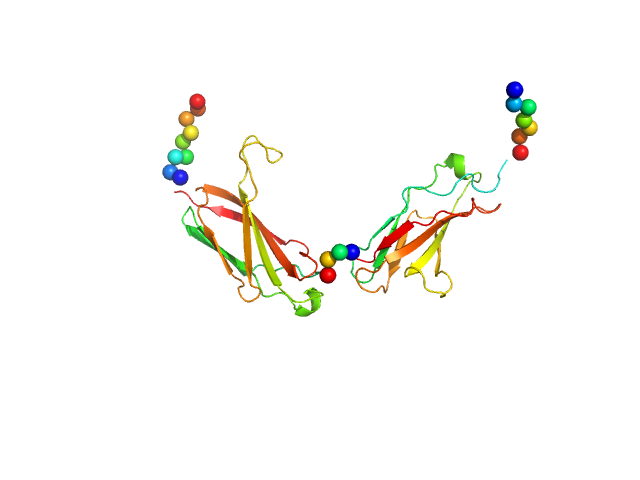
|
| Sample: |
Neural cell adhesion molecule L1 monomer, 24 kDa Homo sapiens protein
|
| Buffer: |
50 mM MES pH 6.5, 150 mM NaCl, 5% glycerol, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 May 11
|
X‐ray structure and function of fibronectin domains two and three of the neural cell adhesion molecule L1
The FASEB Journal 37(3) (2023)
Guédez G, Loers G, Jeffries C, Kozak S, Meijers R, Svergun D, Schachner M, Löw C
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
72 |
nm3 |
|
|
UniProt ID: P0DP23 (1-149) Calmodulin-1
|
|
|
|
| Sample: |
Calmodulin-1 monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 2 mM CaCl2, 1 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Sep 23
|
Binding by calmodulin is coupled to transient unfolding of the third FF domain of Prp40A.
Protein Sci 32(4):e4606 (2023)
Díaz Casas A, Cordoba JJ, Ferrer BJ, Balakrishnan S, Wurm JE, Pastrana-Ríos B, Chazin WJ
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
38 |
nm3 |
|
|