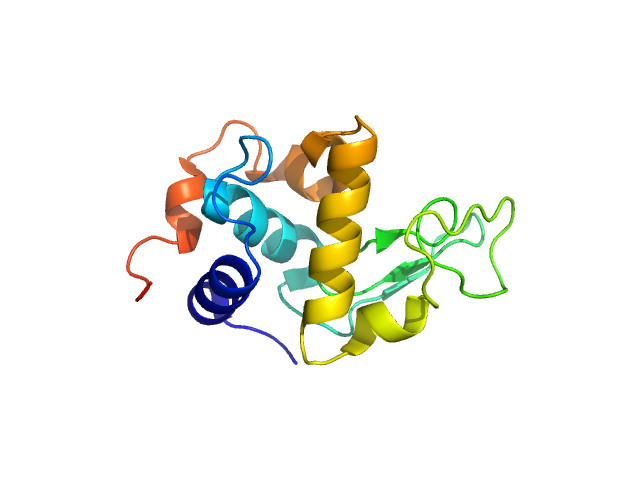
UniProt ID: P00698 (19-147) Lysozyme C
|
|
|
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
50 mM sodium citrate, 150 mM NaCl, pH: 4.5 |
| Experiment: |
SANS
data collected at (Consensus SAS), Multi-facility, Multiple countries on 2022 Jun 22
|
A round-robin approach provides a detailed assessment of biomolecular small-angle scattering data reproducibility and yields consensus curves for benchmarking
Acta Crystallographica Section D Structural Biology 78(11) (2022)
Trewhella J, Vachette P, Bierma J, Blanchet C, Brookes E, Chakravarthy S, Chatzimagas L, Cleveland T, Cowieson N, Crossett B, Duff A, Franke D, Gabel F, Gillilan R, Graewert M, Grishaev A, Guss J, Hammel M, Hopkins J, Huang Q, Hub J, Hura G, Irving T, Jeffries C, Jeong C, Kirby N, Krueger S, Martel A, Matsui T, Li N, Pérez J, Porcar L, Prangé T, Rajkovic I, Rocco M, Rosenberg D, Ryan T, Seifert S, Sekiguchi H, Svergun D, Teixeira S, Thureau A, Weiss T, Whitten A, Wood K, Zuo X
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.8 |
nm |
|
|
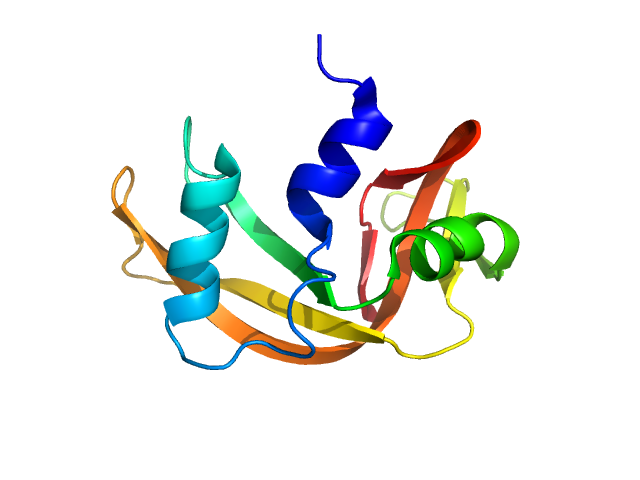
UniProt ID: P61823 (27-150) Ribonuclease pancreatic
|
|
|
|
| Sample: |
Ribonuclease pancreatic monomer, 14 kDa Bos taurus protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SANS
data collected at (Consensus SAS), Multi-facility, Multiple countries on 2022 Jun 22
|
A round-robin approach provides a detailed assessment of biomolecular small-angle scattering data reproducibility and yields consensus curves for benchmarking
Acta Crystallographica Section D Structural Biology 78(11) (2022)
Trewhella J, Vachette P, Bierma J, Blanchet C, Brookes E, Chakravarthy S, Chatzimagas L, Cleveland T, Cowieson N, Crossett B, Duff A, Franke D, Gabel F, Gillilan R, Graewert M, Grishaev A, Guss J, Hammel M, Hopkins J, Huang Q, Hub J, Hura G, Irving T, Jeffries C, Jeong C, Kirby N, Krueger S, Martel A, Matsui T, Li N, Pérez J, Porcar L, Prangé T, Rajkovic I, Rocco M, Rosenberg D, Ryan T, Seifert S, Sekiguchi H, Svergun D, Teixeira S, Thureau A, Weiss T, Whitten A, Wood K, Zuo X
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.1 |
nm |
|
|
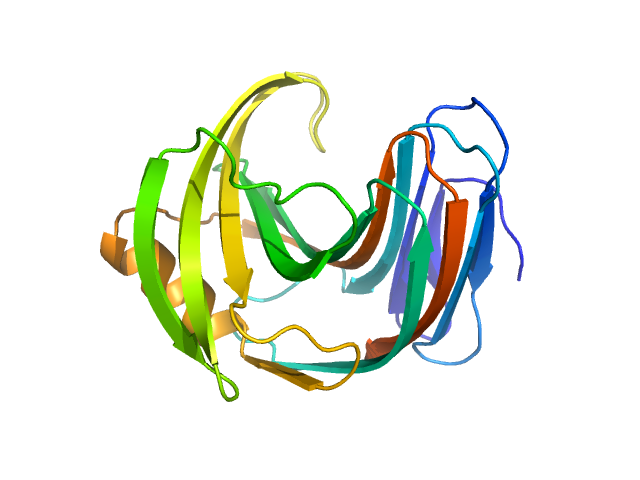
UniProt ID: F8W669 (1-190) Endo-1,4-beta-xylanase
|
|
|
|
| Sample: |
Endo-1,4-beta-xylanase monomer, 21 kDa Trichoderma longibrachiatum protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SANS
data collected at (Consensus SAS), Multi-facility, Multiple countries on 2022 Jun 22
|
A round-robin approach provides a detailed assessment of biomolecular small-angle scattering data reproducibility and yields consensus curves for benchmarking
Acta Crystallographica Section D Structural Biology 78(11) (2022)
Trewhella J, Vachette P, Bierma J, Blanchet C, Brookes E, Chakravarthy S, Chatzimagas L, Cleveland T, Cowieson N, Crossett B, Duff A, Franke D, Gabel F, Gillilan R, Graewert M, Grishaev A, Guss J, Hammel M, Hopkins J, Huang Q, Hub J, Hura G, Irving T, Jeffries C, Jeong C, Kirby N, Krueger S, Martel A, Matsui T, Li N, Pérez J, Porcar L, Prangé T, Rajkovic I, Rocco M, Rosenberg D, Ryan T, Seifert S, Sekiguchi H, Svergun D, Teixeira S, Thureau A, Weiss T, Whitten A, Wood K, Zuo X
|
| RgGuinier |
1.6 |
nm |
| Dmax |
4.3 |
nm |
|
|
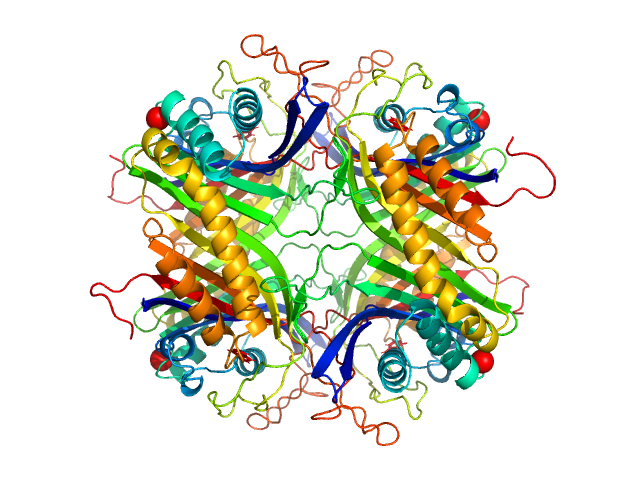
UniProt ID: Q00511 (2-302) Uricase
|
|
|
|
| Sample: |
Uricase tetramer, 136 kDa Aspergillus flavus protein
|
| Buffer: |
100 mM Tris, 150 mM NaCl, pH: 8 |
| Experiment: |
SANS
data collected at (Consensus SAS), Multi-facility, Multiple countries on 2022 Jun 24
|
A round-robin approach provides a detailed assessment of biomolecular small-angle scattering data reproducibility and yields consensus curves for benchmarking
Acta Crystallographica Section D Structural Biology 78(11) (2022)
Trewhella J, Vachette P, Bierma J, Blanchet C, Brookes E, Chakravarthy S, Chatzimagas L, Cleveland T, Cowieson N, Crossett B, Duff A, Franke D, Gabel F, Gillilan R, Graewert M, Grishaev A, Guss J, Hammel M, Hopkins J, Huang Q, Hub J, Hura G, Irving T, Jeffries C, Jeong C, Kirby N, Krueger S, Martel A, Matsui T, Li N, Pérez J, Porcar L, Prangé T, Rajkovic I, Rocco M, Rosenberg D, Ryan T, Seifert S, Sekiguchi H, Svergun D, Teixeira S, Thureau A, Weiss T, Whitten A, Wood K, Zuo X
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.1 |
nm |
|
|
UniProt ID: P24300 (1-388) Xylose isomerase
|
|
|
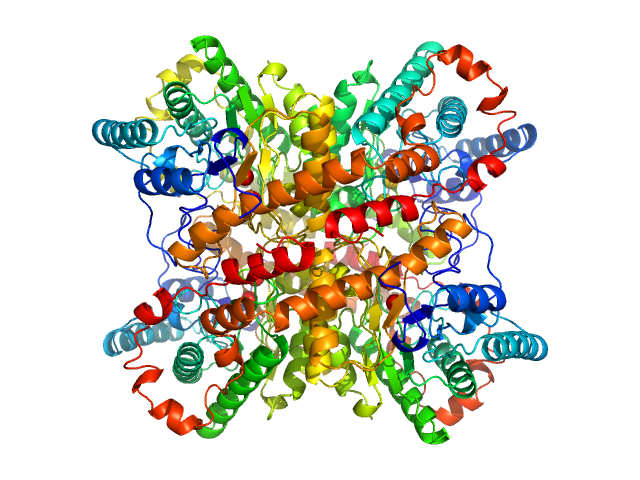
|
| Sample: |
Xylose isomerase tetramer, 173 kDa Streptomyces rubiginosus protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, 1 mM MgCl2, pH: 7.5 |
| Experiment: |
SANS
data collected at (Consensus SAS), Multi-facility, Multiple countries on 2022 Jun 24
|
A round-robin approach provides a detailed assessment of biomolecular small-angle scattering data reproducibility and yields consensus curves for benchmarking
Acta Crystallographica Section D Structural Biology 78(11) (2022)
Trewhella J, Vachette P, Bierma J, Blanchet C, Brookes E, Chakravarthy S, Chatzimagas L, Cleveland T, Cowieson N, Crossett B, Duff A, Franke D, Gabel F, Gillilan R, Graewert M, Grishaev A, Guss J, Hammel M, Hopkins J, Huang Q, Hub J, Hura G, Irving T, Jeffries C, Jeong C, Kirby N, Krueger S, Martel A, Matsui T, Li N, Pérez J, Porcar L, Prangé T, Rajkovic I, Rocco M, Rosenberg D, Ryan T, Seifert S, Sekiguchi H, Svergun D, Teixeira S, Thureau A, Weiss T, Whitten A, Wood K, Zuo X
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.7 |
nm |
|
|
UniProt ID: P03416 (1-454) Nucleoprotein
|
|
|
|
| Sample: |
Nucleoprotein monomer, 50 kDa Murine coronavirus (strain … protein
|
| Buffer: |
70 mM KCl, 50 μM EDTA, 25 mM HEPES, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2021 Jun 10
|
MHV Coronavirus Packaging Signal RNA and N protein
Suzette Pabit
|
| RgGuinier |
5.1 |
nm |
| Dmax |
11.9 |
nm |
| VolumePorod |
91 |
nm3 |
|
|
UniProt ID: Q59QB8 (25-245) Gcf1p
|
|
|
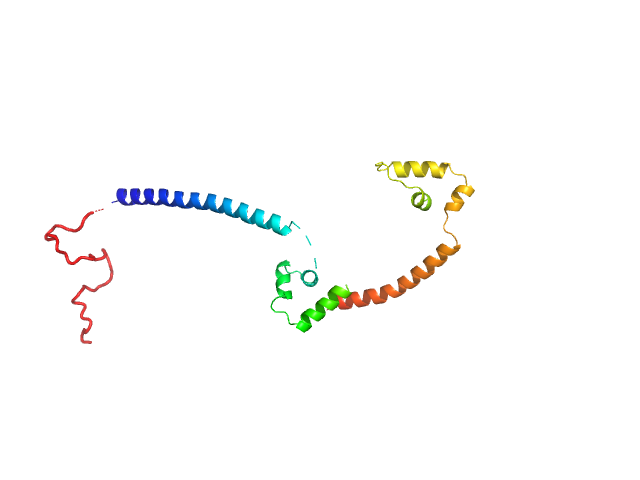
|
| Sample: |
Gcf1p monomer, 26 kDa Candida albicans (strain … protein
|
| Buffer: |
50mM Tris, 750mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Oct 10
|
Structural analysis of the Candida albicans mitochondrial DNA maintenance factor Gcf1p reveals a dynamic DNA-bridging mechanism.
Nucleic Acids Res (2023)
Tarrés-Solé A, Battistini F, Gerhold JM, Piétrement O, Martínez-García B, Ruiz-López E, Lyonnais S, Bernadó P, Roca J, Orozco M, Le Cam E, Sedman J, Solà M
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
80 |
nm3 |
|
|
UniProt ID: Q59QB8 (25-245) Gcf1p
UniProt ID: None (None-None) Af2_20 DNA
|
|
|
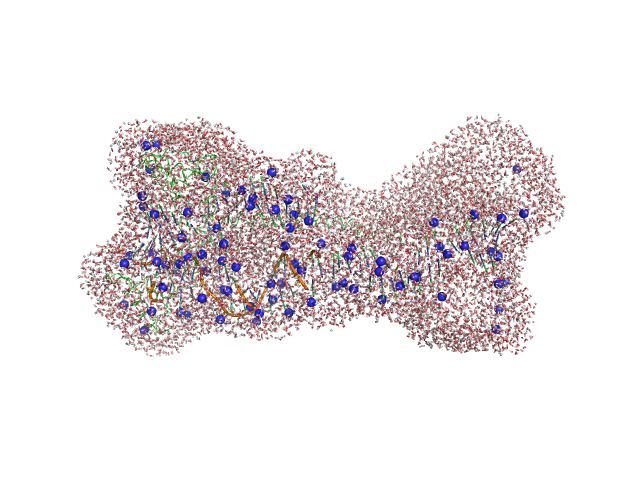
|
| Sample: |
Gcf1p monomer, 26 kDa Candida albicans (strain … protein
Af2_20 DNA monomer, 12 kDa DNA
|
| Buffer: |
25 mM Tris, 20 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Aug 30
|
Structural analysis of the Candida albicans mitochondrial DNA maintenance factor Gcf1p reveals a dynamic DNA-bridging mechanism.
Nucleic Acids Res (2023)
Tarrés-Solé A, Battistini F, Gerhold JM, Piétrement O, Martínez-García B, Ruiz-López E, Lyonnais S, Bernadó P, Roca J, Orozco M, Le Cam E, Sedman J, Solà M
|
| RgGuinier |
3.6 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
UniProt ID: Q59QB8 (25-245) Gcf1p
UniProt ID: None (None-None) Af2_20 DNA
|
|
|
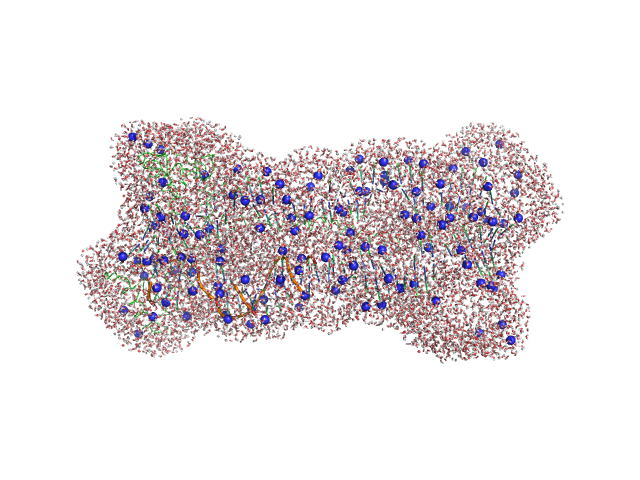
|
| Sample: |
Gcf1p monomer, 26 kDa Candida albicans (strain … protein
Af2_20 DNA monomer, 12 kDa DNA
|
| Buffer: |
25 mM Tris, 20 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Aug 30
|
Structural analysis of the Candida albicans mitochondrial DNA maintenance factor Gcf1p reveals a dynamic DNA-bridging mechanism.
Nucleic Acids Res (2023)
Tarrés-Solé A, Battistini F, Gerhold JM, Piétrement O, Martínez-García B, Ruiz-López E, Lyonnais S, Bernadó P, Roca J, Orozco M, Le Cam E, Sedman J, Solà M
|
| RgGuinier |
3.9 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
133 |
nm3 |
|
|
UniProt ID: None (None-None) Af2_20 DNA
UniProt ID: Q59QB8 (59-245) Gcf1p(Δ58)
|
|
|
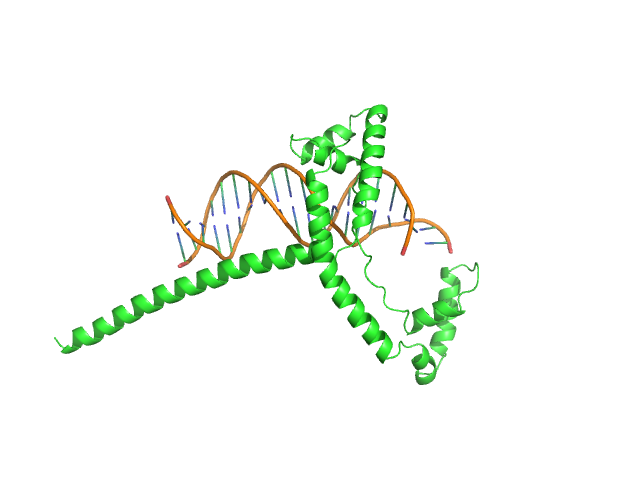
|
| Sample: |
Af2_20 DNA monomer, 12 kDa DNA
Gcf1p(Δ58) monomer, 22 kDa Candida albicans (strain … protein
|
| Buffer: |
25 mM Tris, 20 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Aug 30
|
Structural analysis of the Candida albicans mitochondrial DNA maintenance factor Gcf1p reveals a dynamic DNA-bridging mechanism.
Nucleic Acids Res (2023)
Tarrés-Solé A, Battistini F, Gerhold JM, Piétrement O, Martínez-García B, Ruiz-López E, Lyonnais S, Bernadó P, Roca J, Orozco M, Le Cam E, Sedman J, Solà M
|
| RgGuinier |
2.6 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
52 |
nm3 |
|
|