UniProt ID: B2V8L9 (2-496) SAVED domain-containing protein
|
|
|

|
| Sample: |
SAVED domain-containing protein monomer, 58 kDa Sulfurihydrogenibium sp. (strain … protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Apr 8
|
Antiviral signalling by a cyclic nucleotide activated CRISPR protease.
Nature 614(7946):168-174 (2023)
Rouillon C, Schneberger N, Chi H, Blumenstock K, Da Vela S, Ackermann K, Moecking J, Peter MF, Boenigk W, Seifert R, Bode BE, Schmid-Burgk JL, Svergun D, Geyer M, White MF, Hagelueken G
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
116 |
nm3 |
|
|
UniProt ID: A0A100HLL5 (10-214) DNA protection during starvation, DPS (Ferritin superfamily)
|
|
|

|
| Sample: |
DNA protection during starvation, DPS (Ferritin superfamily) dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 230 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 8
|
Controlled modulation of the dynamics of the Deinococcus grandis Dps N-terminal tails by divalent metals.
Protein Sci :e4567 (2023)
Guerra JPL, Blanchet CE, Vieira BJC, Waerenborgh JC, Jones NC, Hoffmann SV, Pereira AS, Tavares P
|
| RgGuinier |
4.5 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
454 |
nm3 |
|
|
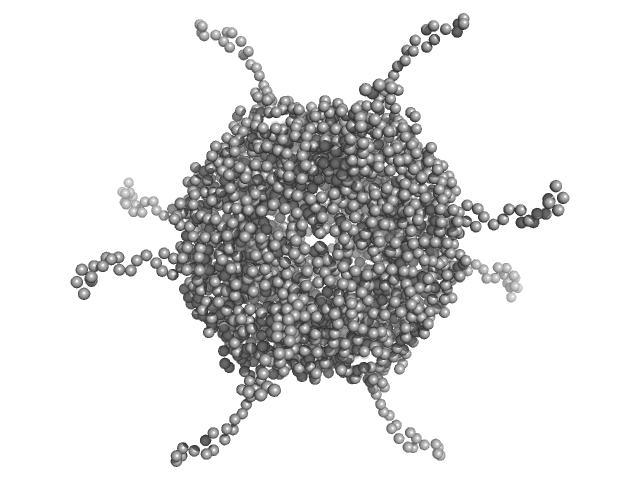
UniProt ID: A0A100HLL5 (10-214) DNA protection during starvation, DPS (Ferritin superfamily)
|
|
|
|
| Sample: |
DNA protection during starvation, DPS (Ferritin superfamily) dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 230 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 8
|
Controlled modulation of the dynamics of the Deinococcus grandis Dps N-terminal tails by divalent metals.
Protein Sci :e4567 (2023)
Guerra JPL, Blanchet CE, Vieira BJC, Waerenborgh JC, Jones NC, Hoffmann SV, Pereira AS, Tavares P
|
| RgGuinier |
4.4 |
nm |
| Dmax |
18.4 |
nm |
| VolumePorod |
440 |
nm3 |
|
|
UniProt ID: A0A100HLL5 (10-214) DNA protection during starvation, DPS (Ferritin superfamily)
|
|
|

|
| Sample: |
DNA protection during starvation, DPS (Ferritin superfamily) dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 230 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 8
|
Controlled modulation of the dynamics of the Deinococcus grandis Dps N-terminal tails by divalent metals.
Protein Sci :e4567 (2023)
Guerra JPL, Blanchet CE, Vieira BJC, Waerenborgh JC, Jones NC, Hoffmann SV, Pereira AS, Tavares P
|
| RgGuinier |
4.2 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
464 |
nm3 |
|
|
UniProt ID: A0A100HLL5 (10-214) DNA protection during starvation, DPS (Ferritin superfamily)
|
|
|

|
| Sample: |
DNA protection during starvation, DPS (Ferritin superfamily) dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 230 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 8
|
Controlled modulation of the dynamics of the Deinococcus grandis Dps N-terminal tails by divalent metals.
Protein Sci :e4567 (2023)
Guerra JPL, Blanchet CE, Vieira BJC, Waerenborgh JC, Jones NC, Hoffmann SV, Pereira AS, Tavares P
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
439 |
nm3 |
|
|
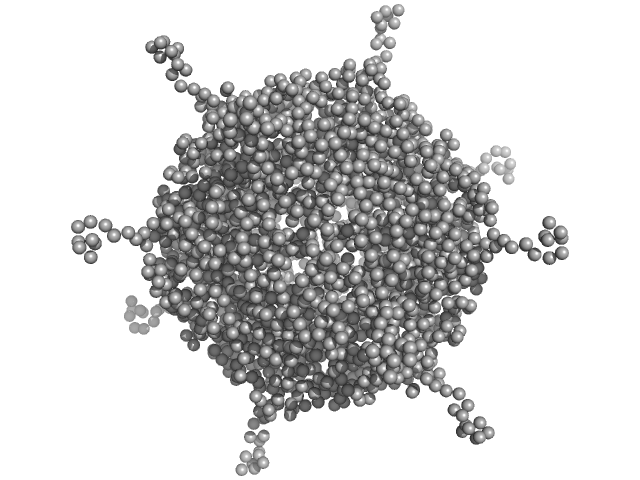
UniProt ID: A0A100HLL5 (10-214) DNA protection during starvation, DPS (Ferritin superfamily)
|
|
|
|
| Sample: |
DNA protection during starvation, DPS (Ferritin superfamily) dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 230 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 8
|
Controlled modulation of the dynamics of the Deinococcus grandis Dps N-terminal tails by divalent metals.
Protein Sci :e4567 (2023)
Guerra JPL, Blanchet CE, Vieira BJC, Waerenborgh JC, Jones NC, Hoffmann SV, Pereira AS, Tavares P
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
427 |
nm3 |
|
|
UniProt ID: A0A100HLL5 (10-214) DNA protection during starvation, DPS (Ferritin superfamily)
|
|
|

|
| Sample: |
DNA protection during starvation, DPS (Ferritin superfamily) dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS, 230 mM NaCl, 5 mM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 8
|
Controlled modulation of the dynamics of the Deinococcus grandis Dps N-terminal tails by divalent metals.
Protein Sci :e4567 (2023)
Guerra JPL, Blanchet CE, Vieira BJC, Waerenborgh JC, Jones NC, Hoffmann SV, Pereira AS, Tavares P
|
| RgGuinier |
4.5 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
453 |
nm3 |
|
|
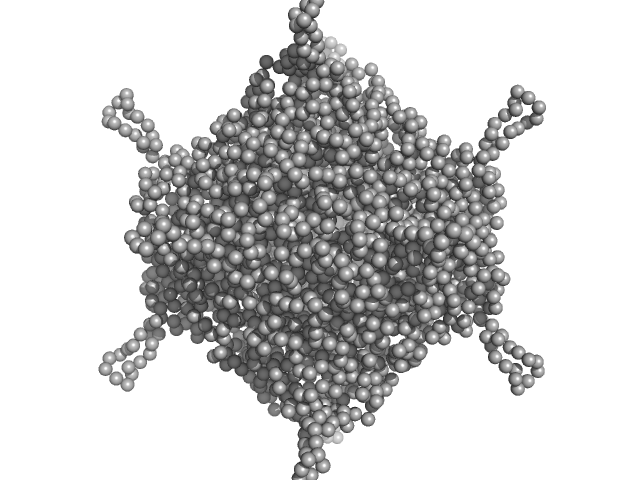
UniProt ID: A0A100HLL5 (10-214) DNA protection during starvation, DPS (Ferritin superfamily)
|
|
|
|
| Sample: |
DNA protection during starvation, DPS (Ferritin superfamily) dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 230 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 8
|
Controlled modulation of the dynamics of the Deinococcus grandis Dps N-terminal tails by divalent metals.
Protein Sci :e4567 (2023)
Guerra JPL, Blanchet CE, Vieira BJC, Waerenborgh JC, Jones NC, Hoffmann SV, Pereira AS, Tavares P
|
| RgGuinier |
4.2 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
458 |
nm3 |
|
|
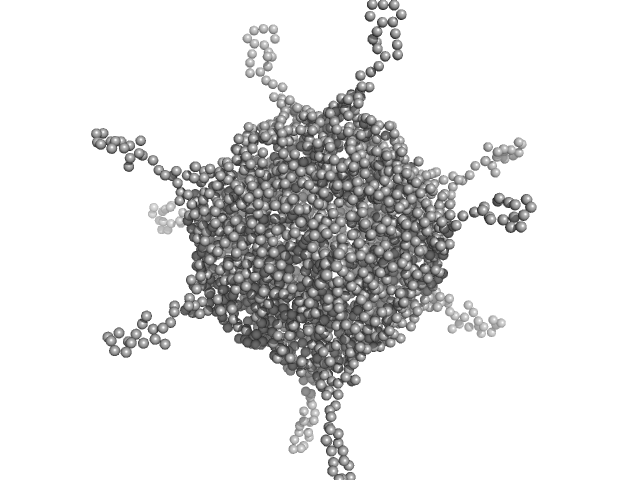
UniProt ID: A0A100HLL5 (10-214) DNA protection during starvation, DPS (Ferritin superfamily)
|
|
|
|
| Sample: |
DNA protection during starvation, DPS (Ferritin superfamily) dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 230 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 8
|
Controlled modulation of the dynamics of the Deinococcus grandis Dps N-terminal tails by divalent metals.
Protein Sci :e4567 (2023)
Guerra JPL, Blanchet CE, Vieira BJC, Waerenborgh JC, Jones NC, Hoffmann SV, Pereira AS, Tavares P
|
| RgGuinier |
4.4 |
nm |
| Dmax |
19.3 |
nm |
| VolumePorod |
438 |
nm3 |
|
|
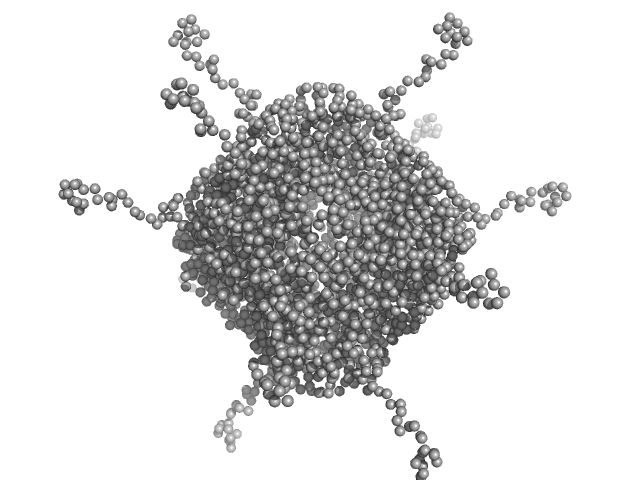
UniProt ID: A0A100HLL5 (11-214) DNA protection during starvation, DPS-D43A (Ferritin superfamily) dodecamer D43A mutation
|
|
|
|
| Sample: |
DNA protection during starvation, DPS-D43A (Ferritin superfamily) dodecamer D43A mutation dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 230 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 8
|
Controlled modulation of the dynamics of the Deinococcus grandis Dps N-terminal tails by divalent metals.
Protein Sci :e4567 (2023)
Guerra JPL, Blanchet CE, Vieira BJC, Waerenborgh JC, Jones NC, Hoffmann SV, Pereira AS, Tavares P
|
| RgGuinier |
4.5 |
nm |
| Dmax |
19.8 |
nm |
| VolumePorod |
451 |
nm3 |
|
|