UniProt ID: Q13363 (1-440) C-terminal-binding protein 1
UniProt ID: Q9Y5P3 (303-465) Retinoic acid-induced protein 2 (303-465: L345A, S346A)
|
|
|
|
| Sample: |
C-terminal-binding protein 1 tetramer, 191 kDa Homo sapiens protein
Retinoic acid-induced protein 2 (303-465: L345A, S346A) monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 1 mM TCEP, 5% v/v glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Apr 13
|
Master corepressor inactivation through multivalent SLiM-induced polymerization mediated by the oncogene suppressor RAI2.
Nat Commun 15(1):5241 (2024)
Goradia N, Werner S, Mullapudi E, Greimeier S, Bergmann L, Lang A, Mertens H, Węglarz A, Sander S, Chojnowski G, Wikman H, Ohlenschläger O, von Amsberg G, Pantel K, Wilmanns M
|
| RgGuinier |
4.8 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
310 |
nm3 |
|
|
UniProt ID: Q13363 (1-440) C-terminal-binding protein 1
UniProt ID: Q9Y5P3 (303-465) Retinoic acid-induced protein 2 (303-465: L319A, S320A, L345A, S346A)
|
|
|
|
| Sample: |
C-terminal-binding protein 1 tetramer, 191 kDa Homo sapiens protein
Retinoic acid-induced protein 2 (303-465: L319A, S320A, L345A, S346A) monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 1 mM TCEP, 5% v/v glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Apr 13
|
Master corepressor inactivation through multivalent SLiM-induced polymerization mediated by the oncogene suppressor RAI2.
Nat Commun 15(1):5241 (2024)
Goradia N, Werner S, Mullapudi E, Greimeier S, Bergmann L, Lang A, Mertens H, Węglarz A, Sander S, Chojnowski G, Wikman H, Ohlenschläger O, von Amsberg G, Pantel K, Wilmanns M
|
| RgGuinier |
5.1 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
387 |
nm3 |
|
|
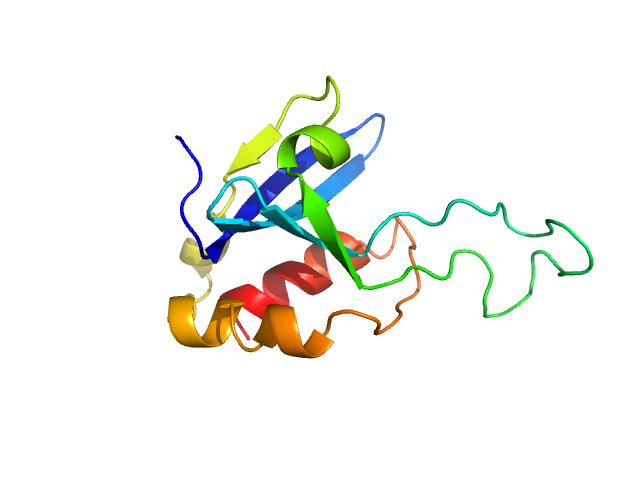
UniProt ID: Q9UL19 (1-123) Phospholipase A and acyltransferase 4
|
|
|
|
| Sample: |
Phospholipase A and acyltransferase 4 monomer, 14 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM TECP, 1% Glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Mar 11
|
Crystal structure of the phospholipase A and acyltransferase 4 (PLAAT4) catalytic domain.
J Struct Biol 214(4):107903 (2022)
Wehlin A, Cornaciu I, Marquez JA, Perrakis A, von Castelmur E
|
| RgGuinier |
1.9 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
31 |
nm3 |
|
|
UniProt ID: P60842 (1-406) Eukaryotic initiation factor 4A-I
|
|
|
|
| Sample: |
Eukaryotic initiation factor 4A-I monomer, 46 kDa Homo sapiens protein
|
| Buffer: |
20 mM Hepes, 100 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Jul 22
|
eIF4A1-dependent mRNAs employ purine-rich 5'UTR sequences to activate localised eIF4A1-unwinding through eIF4A1-multimerisation to facilitate translation.
Nucleic Acids Res (2023)
Schmidt T, Dabrowska A, Waldron JA, Hodge K, Koulouras G, Gabrielsen M, Munro J, Tack DC, Harris G, McGhee E, Scott D, Carlin LM, Huang D, Le Quesne J, Zanivan S, Wilczynska A, Bushell M
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
80 |
nm3 |
|
|
UniProt ID: P60842 (1-406) Eukaryotic initiation factor 4A-I
UniProt ID: None (None-None) (AG)10-RNA
|
|
|
|
| Sample: |
Eukaryotic initiation factor 4A-I monomer, 46 kDa Homo sapiens protein
(AG)10-RNA monomer, 7 kDa synthetic construct RNA
|
| Buffer: |
20 mM Hepes, 100 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Sep 16
|
eIF4A1-dependent mRNAs employ purine-rich 5'UTR sequences to activate localised eIF4A1-unwinding through eIF4A1-multimerisation to facilitate translation.
Nucleic Acids Res (2023)
Schmidt T, Dabrowska A, Waldron JA, Hodge K, Koulouras G, Gabrielsen M, Munro J, Tack DC, Harris G, McGhee E, Scott D, Carlin LM, Huang D, Le Quesne J, Zanivan S, Wilczynska A, Bushell M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
96 |
nm3 |
|
|
UniProt ID: P60842 (1-406) Eukaryotic initiation factor 4A-I
UniProt ID: None (None-None) (CAA)6CA-RNA
|
|
|
|
| Sample: |
Eukaryotic initiation factor 4A-I monomer, 46 kDa Homo sapiens protein
(CAA)6CA-RNA monomer, 6 kDa synthetic construct RNA
|
| Buffer: |
20 mM Hepes, 100 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Jul 22
|
eIF4A1-dependent mRNAs employ purine-rich 5'UTR sequences to activate localised eIF4A1-unwinding through eIF4A1-multimerisation to facilitate translation.
Nucleic Acids Res (2023)
Schmidt T, Dabrowska A, Waldron JA, Hodge K, Koulouras G, Gabrielsen M, Munro J, Tack DC, Harris G, McGhee E, Scott D, Carlin LM, Huang D, Le Quesne J, Zanivan S, Wilczynska A, Bushell M
|
| RgGuinier |
3.1 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
UniProt ID: None (None-None) (AG)10-RNA
UniProt ID: P60842 (1-406) Eukaryotic initiation factor 4A-I
|
|
|
|
| Sample: |
(AG)10-RNA monomer, 7 kDa synthetic construct RNA
Eukaryotic initiation factor 4A-I trimer, 138 kDa Homo sapiens protein
|
| Buffer: |
20 mM Hepes, 100 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Sep 16
|
eIF4A1-dependent mRNAs employ purine-rich 5'UTR sequences to activate localised eIF4A1-unwinding through eIF4A1-multimerisation to facilitate translation.
Nucleic Acids Res (2023)
Schmidt T, Dabrowska A, Waldron JA, Hodge K, Koulouras G, Gabrielsen M, Munro J, Tack DC, Harris G, McGhee E, Scott D, Carlin LM, Huang D, Le Quesne J, Zanivan S, Wilczynska A, Bushell M
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.1 |
nm |
| VolumePorod |
150 |
nm3 |
|
|
UniProt ID: P60842 (1-406) Eukaryotic initiation factor 4A-I
UniProt ID: None (None-None) (AG)10-24bp loading-RNA
UniProt ID: None (None-None) (AG)10-24bp duplex-RNA
|
|
|
|
| Sample: |
Eukaryotic initiation factor 4A-I trimer, 138 kDa Homo sapiens protein
(AG)10-24bp loading-RNA monomer, 15 kDa synthetic construct RNA
(AG)10-24bp duplex-RNA monomer, 7 kDa synthetic construct RNA
|
| Buffer: |
20 mM Hepes, 100 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Sep 16
|
eIF4A1-dependent mRNAs employ purine-rich 5'UTR sequences to activate localised eIF4A1-unwinding through eIF4A1-multimerisation to facilitate translation.
Nucleic Acids Res (2023)
Schmidt T, Dabrowska A, Waldron JA, Hodge K, Koulouras G, Gabrielsen M, Munro J, Tack DC, Harris G, McGhee E, Scott D, Carlin LM, Huang D, Le Quesne J, Zanivan S, Wilczynska A, Bushell M
|
| RgGuinier |
4.9 |
nm |
| Dmax |
16.2 |
nm |
| VolumePorod |
230 |
nm3 |
|
|
UniProt ID: Q0P7K1 (1-2154) Colibactin hybrid non-ribosomal peptide synthetase/type I polyketide synthase ClbK
|
|
|
|
| Sample: |
Colibactin hybrid non-ribosomal peptide synthetase/type I polyketide synthase ClbK dimer, 476 kDa Escherichia coli protein
|
| Buffer: |
50 mM Hepes pH 7.5, 200 mM NaCl, 5 mM DTT, 5 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Sep 14
|
Architecture of a PKS-NRPS hybrid megaenzyme involved in the biosynthesis of the genotoxin colibactin
Structure (2023)
Bonhomme S, Contreras-Martel C, Dessen A, Macheboeuf P
|
| RgGuinier |
8.0 |
nm |
| Dmax |
30.0 |
nm |
| VolumePorod |
1340 |
nm3 |
|
|
UniProt ID: Q06609 (1-339) DNA repair protein RAD51 homolog 1 (F86E A89E)
|
|
|
|
| Sample: |
DNA repair protein RAD51 homolog 1 (F86E A89E) monomer, 37 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 200 mM Na2SO4, 5% glycerol, 0.1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Apr 7
|
Isolation and characterization of monomeric human RAD51: a novel tool for investigating homologous recombination in cancer
Angewandte Chemie International Edition (2023)
Rinaldi F, Schipani F, Balboni B, Catalano F, Marotta R, Myers S, Previtali V, Veronesi M, Scietti L, Cecatiello V, Pasqualato S, Ortega J, Girotto S, Cavalli A
|
| RgGuinier |
3.0 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
61 |
nm3 |
|
|