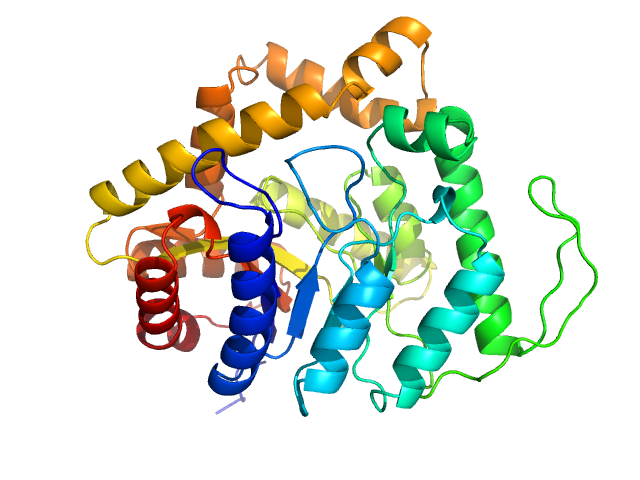
UniProt ID: A0A2S9XZH0 (1-363) Alkanal monooxygenase alpha chain
|
|
|
|
| Sample: |
Alkanal monooxygenase alpha chain dimer, 84 kDa Enhygromyxa salina protein
|
| Buffer: |
150 mM NaCl, 25 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2020 Aug 1
|
luxA
Gene From Enhygromyxa salina
Encodes a Functional Homodimeric Luciferase
Proteins: Structure, Function, and Bioinformatics (2024)
Yudenko A, Bazhenov S, Aleksenko V, Goncharov I, Semenov O, Remeeva A, Nazarenko V, Kuznetsova E, Fomin V, Konopleva M, Al Ebrahim R, Sluchanko N, Ryzhykau Y, Semenov Y, Kuklin A, Manukhov I, Gushchin I
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
138 |
nm3 |
|
|
UniProt ID: O55777 (200-310) Non-structural protein V
|
|
|
|
| Sample: |
Non-structural protein V monomer, 15 kDa Hendra virus (isolate … protein
|
| Buffer: |
50 mM sodium phosphate, pH: 7.2 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Jul 14
|
Molecular Determinants of Fibrillation in a Viral Amyloidogenic Domain from Combined Biochemical and Biophysical Studies
International Journal of Molecular Sciences 24(1):399 (2022)
Nilsson J, Baroudi H, Gondelaud F, Pesce G, Bignon C, Ptchelkine D, Chamieh J, Cottet H, Kajava A, Longhi S
|
| RgGuinier |
3.7 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
UniProt ID: O55777 (200-310) Non-structural protein V (Y211A, Y212A, Y213A mutant)
|
|
|
|
| Sample: |
Non-structural protein V (Y211A, Y212A, Y213A mutant) monomer, 15 kDa Hendra virus (isolate … protein
|
| Buffer: |
50 mM sodium phosphate, pH: 7.2 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Jul 14
|
Molecular Determinants of Fibrillation in a Viral Amyloidogenic Domain from Combined Biochemical and Biophysical Studies
International Journal of Molecular Sciences 24(1):399 (2022)
Nilsson J, Baroudi H, Gondelaud F, Pesce G, Bignon C, Ptchelkine D, Chamieh J, Cottet H, Kajava A, Longhi S
|
| RgGuinier |
4.0 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
UniProt ID: O55777 (200-254) Non-structural protein V
|
|
|
|
| Sample: |
Non-structural protein V monomer, 9 kDa Hendra virus (isolate … protein
|
| Buffer: |
50 mM sodium phosphate, pH: 7.2 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Jul 14
|
Molecular Determinants of Fibrillation in a Viral Amyloidogenic Domain from Combined Biochemical and Biophysical Studies
International Journal of Molecular Sciences 24(1):399 (2022)
Nilsson J, Baroudi H, Gondelaud F, Pesce G, Bignon C, Ptchelkine D, Chamieh J, Cottet H, Kajava A, Longhi S
|
| RgGuinier |
2.8 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
UniProt ID: O55777 (200-254) Non-structural protein V (ΔC-terminal and Y111A, Y112A, Y113A mutant)
|
|
|
|
| Sample: |
Non-structural protein V (ΔC-terminal and Y111A, Y112A, Y113A mutant) monomer, 9 kDa Hendra virus (isolate … protein
|
| Buffer: |
50 mM sodium phosphate, pH: 7.2 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Jul 14
|
Molecular Determinants of Fibrillation in a Viral Amyloidogenic Domain from Combined Biochemical and Biophysical Studies
International Journal of Molecular Sciences 24(1):399 (2022)
Nilsson J, Baroudi H, Gondelaud F, Pesce G, Bignon C, Ptchelkine D, Chamieh J, Cottet H, Kajava A, Longhi S
|
| RgGuinier |
2.7 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
UniProt ID: Q997F2 (200-314) Non-structural protein V
|
|
|
|
| Sample: |
Non-structural protein V monomer, 15 kDa Nipah henipavirus protein
|
| Buffer: |
50 mM sodium phosphate, pH: 7.2 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Jul 14
|
Molecular Determinants of Fibrillation in a Viral Amyloidogenic Domain from Combined Biochemical and Biophysical Studies
International Journal of Molecular Sciences 24(1):399 (2022)
Nilsson J, Baroudi H, Gondelaud F, Pesce G, Bignon C, Ptchelkine D, Chamieh J, Cottet H, Kajava A, Longhi S
|
| RgGuinier |
3.7 |
nm |
| Dmax |
16.5 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
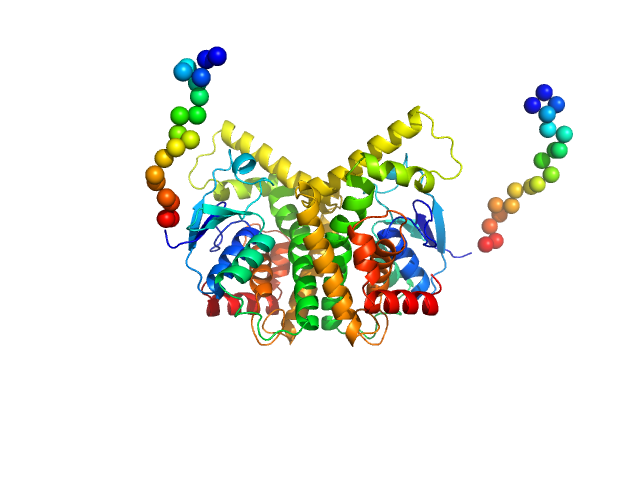
UniProt ID: A0A222TSG8 (1-238) GST N-terminal domain-containing protein
|
|
|
|
| Sample: |
GST N-terminal domain-containing protein dimer, 60 kDa Gordonia rubripertincta protein
|
| Buffer: |
10 mM Tris, 300 mM NaCl,, pH: 8 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 Q-Xoom, Center for Structural Studies, Heinrich-Heine-University on 2022 Jan 20
|
Glutathione S
-Transferase Mediated Epoxide Conversion: Functional and Structural Properties of an Enantioselective Catalyst
ACS Catalysis :12308-12324 (2025)
Haarmann M, Scholz M, Lienkamp A, Burnik J, Justesen B, Reiners J, Maier A, Eggerichs D, Vocke K, Smits S, Günther Pomorski T, Hofmann E, Tischler D
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
91 |
nm3 |
|
|
UniProt ID: P54578 (1-494) Ubiquitin carboxyl-terminal hydrolase 14
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDQZ7_fit2_model1.png)
|
| Sample: |
Ubiquitin carboxyl-terminal hydrolase 14 monomer, 56 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Oct 18
|
Transient interdomain interactions in free USP14 shape its conformational ensemble.
Protein Sci 33(5):e4975 (2024)
Salomonsson J, Wallner B, Sjöstrand L, D'Arcy P, Sunnerhagen M, Ahlner A
|
| RgGuinier |
3.1 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
111 |
nm3 |
|
|
UniProt ID: P54578 (91-494) Ubiquitin carboxyl-terminal hydrolase 14
|
|
|

|
| Sample: |
Ubiquitin carboxyl-terminal hydrolase 14 monomer, 46 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 5
|
Transient interdomain interactions in free USP14 shape its conformational ensemble.
Protein Sci 33(5):e4975 (2024)
Salomonsson J, Wallner B, Sjöstrand L, D'Arcy P, Sunnerhagen M, Ahlner A
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
88 |
nm3 |
|
|
UniProt ID: P96379 (None-None) Nucleoside triphosphate pyrophosphohydrolase
|
|
|

|
| Sample: |
Nucleoside triphosphate pyrophosphohydrolase dimer, 71 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2022 Sep 4
|
Structural analysis of the housecleaning nucleoside triphosphate pyrophosphohydrolase MazG from Mycobacterium tuberculosis
Frontiers in Microbiology 14 (2023)
Wang S, Gao B, Chen A, Zhang Z, Wang S, Lv L, Zhao G, Li J
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
121 |
nm3 |
|
|