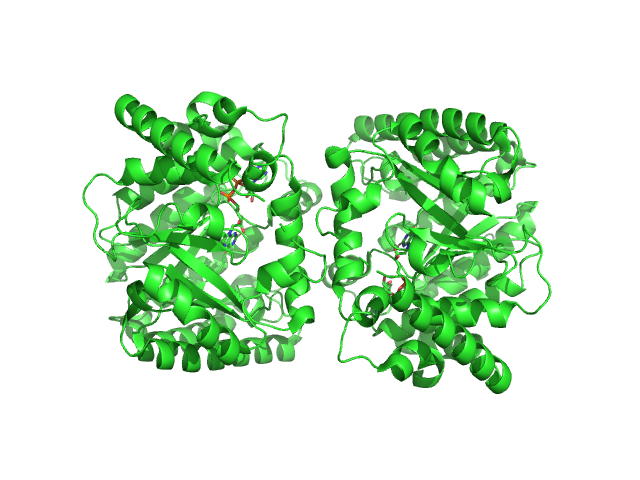
UniProt ID: F7X6I3 (None-None) Minimal proline dehydrogenase domain of proline utilization A (design #2)
|
|
|
|
| Sample: |
Minimal proline dehydrogenase domain of proline utilization A (design #2) dimer, 87 kDa Sinorhizobium meliloti protein
|
| Buffer: |
25 mM HEPES pH 7.6, 150 mM NaCl, and 1mM TCEP, pH: 7.6 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2022 Apr 12
|
Structure-based engineering of minimal Proline dehydrogenase domains for inhibitor discovery.
Protein Eng Des Sel (2022)
Bogner AN, Ji J, Tanner JJ
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
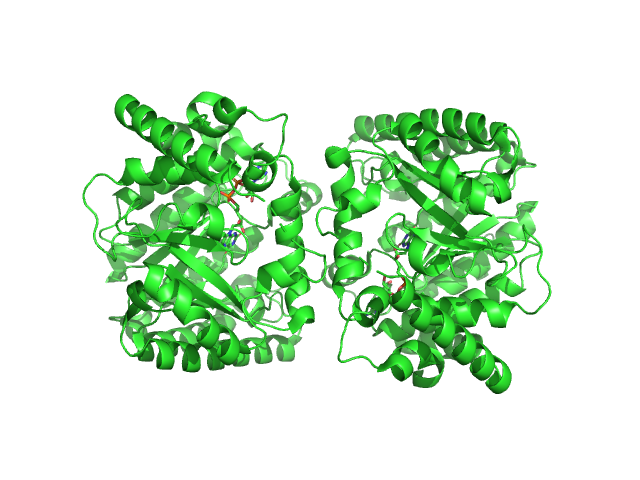
UniProt ID: F7X6I3 (None-None) Minimal proline dehydrogenase domain of proline utilization A (design #2)
|
|
|
|
| Sample: |
Minimal proline dehydrogenase domain of proline utilization A (design #2) dimer, 87 kDa Sinorhizobium meliloti protein
|
| Buffer: |
25 mM HEPES pH 7.6, 150 mM NaCl, and 1mM TCEP, pH: 7.6 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2022 Apr 12
|
Structure-based engineering of minimal Proline dehydrogenase domains for inhibitor discovery.
Protein Eng Des Sel (2022)
Bogner AN, Ji J, Tanner JJ
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
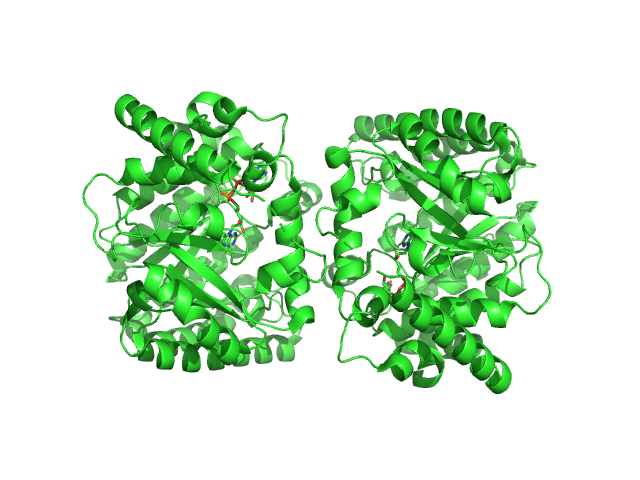
UniProt ID: F7X6I3 (None-None) Minimal proline dehydrogenase domain of proline utilization A (design #2)
|
|
|
|
| Sample: |
Minimal proline dehydrogenase domain of proline utilization A (design #2) dimer, 87 kDa Sinorhizobium meliloti protein
|
| Buffer: |
25 mM HEPES pH 7.6, 150 mM NaCl, and 1mM TCEP, pH: 7.6 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2022 Apr 12
|
Structure-based engineering of minimal Proline dehydrogenase domains for inhibitor discovery.
Protein Eng Des Sel (2022)
Bogner AN, Ji J, Tanner JJ
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
108 |
nm3 |
|
|
UniProt ID: P30177 (1-320) Uncharacterized protein YbiB
UniProt ID: B1XHF9 (1-390) GTPase Obg
|
|
|
|
| Sample: |
Uncharacterized protein YbiB dimer, 75 kDa Escherichia coli (strain … protein
GTPase Obg dimer, 91 kDa Escherichia coli (strain … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM MgCl2, 2 mM DTT, 5% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Oct 24
|
YbiB: a novel interactor of the GTPase ObgE.
Nucleic Acids Res (2023)
Deckers B, Vercauteren S, De Kock V, Martin C, Lazar T, Herpels P, Dewachter L, Verstraeten N, Peeters E, Ballet S, Michiels J, Galicia C, Versées W
|
| RgGuinier |
4.7 |
nm |
| Dmax |
18.6 |
nm |
| VolumePorod |
291 |
nm3 |
|
|
UniProt ID: F8WJV6 (19-1128) p123 expressed protein
|
|
|

|
| Sample: |
P123 expressed protein dimer, 248 kDa Mycoplasma mobile (strain … protein
|
| Buffer: |
100 mM ammonium acetate, pH: 6.8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2019 Oct 25
|
Structure and Function of Gli123 Involved in Mycoplasma mobile Gliding.
J Bacteriol :e0034022 (2023)
Matsuike D, Tahara YO, Nonaka T, Wu HN, Hamaguchi T, Kudo H, Hayashi Y, Arai M, Miyata M
|
| RgGuinier |
8.2 |
nm |
| Dmax |
31.6 |
nm |
| VolumePorod |
436 |
nm3 |
|
|
UniProt ID: Q9BVM2 (1-203) Protein DPCD
|
|
|
|
| Sample: |
Protein DPCD monomer, 23 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris-HCl, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2018 Nov 8
|
DPCD is a regulator of R2TP in ciliogenesis initiation through Akt signaling.
Cell Rep 43(2):113713 (2024)
Mao YQ, Seraphim TV, Wan Y, Wu R, Coyaud E, Bin Munim M, Mollica A, Laurent E, Babu M, Mennella V, Raught B, Houry WA
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
UniProt ID: O95398 (194-923) Rap guanine nucleotide exchange factor 3
|
|
|
|
| Sample: |
Rap guanine nucleotide exchange factor 3 monomer, 82 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM NaCl, 0.5 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2017 Oct 4
|
Membranes prime the RapGEF EPAC1 to transduce cAMP signaling
Nature Communications 14(1) (2023)
Sartre C, Peurois F, Ley M, Kryszke M, Zhang W, Courilleau D, Fischmeister R, Ambroise Y, Zeghouf M, Cianferani S, Ferrandez Y, Cherfils J
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
130 |
nm3 |
|
|
UniProt ID: O95398 (194-923) Rap guanine nucleotide exchange factor 3
|
|
|
|
| Sample: |
Rap guanine nucleotide exchange factor 3 monomer, 82 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM NaCl, 0.5 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2017 Oct 4
|
Membranes prime the RapGEF EPAC1 to transduce cAMP signaling
Nature Communications 14(1) (2023)
Sartre C, Peurois F, Ley M, Kryszke M, Zhang W, Courilleau D, Fischmeister R, Ambroise Y, Zeghouf M, Cianferani S, Ferrandez Y, Cherfils J
|
| RgGuinier |
3.5 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
UniProt ID: O95398 (194-923) Rap guanine nucleotide exchange factor 3
|
|
|
|
| Sample: |
Rap guanine nucleotide exchange factor 3 monomer, 82 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM NaCl, 0.5 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2017 Oct 4
|
Membranes prime the RapGEF EPAC1 to transduce cAMP signaling
Nature Communications 14(1) (2023)
Sartre C, Peurois F, Ley M, Kryszke M, Zhang W, Courilleau D, Fischmeister R, Ambroise Y, Zeghouf M, Cianferani S, Ferrandez Y, Cherfils J
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
UniProt ID: O95398 (194-923) Rap guanine nucleotide exchange factor 3
|
|
|
|
| Sample: |
Rap guanine nucleotide exchange factor 3 monomer, 82 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM NaCl, 0.5 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2017 Oct 4
|
Membranes prime the RapGEF EPAC1 to transduce cAMP signaling
Nature Communications 14(1) (2023)
Sartre C, Peurois F, Ley M, Kryszke M, Zhang W, Courilleau D, Fischmeister R, Ambroise Y, Zeghouf M, Cianferani S, Ferrandez Y, Cherfils J
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
134 |
nm3 |
|
|