UniProt ID: P43351 (1-418) DNA repair protein RAD52 homolog
|
|
|
|
| Sample: |
DNA repair protein RAD52 homolog undecamer, 528 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 7.5, 250 mM NaCl, 1% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Jul 21
|
An integrative structural study of the human full-length RAD52 at 2.2 Å resolution
Communications Biology 7(1) (2024)
Balboni B, Marotta R, Rinaldi F, Milordini G, Varignani G, Girotto S, Cavalli A
|
| RgGuinier |
8.0 |
nm |
| Dmax |
40.2 |
nm |
| VolumePorod |
1218 |
nm3 |
|
|
UniProt ID: A0A6M7H0F8 (2-141) YdaT_toxin domain-containing protein
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDQB9_fit1_model1.png)
|
| Sample: |
YdaT_toxin domain-containing protein tetramer, 74 kDa Escherichia coli O157:H7 protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Apr 16
|
Structural basis of DNA binding by YdaT, a functional equivalent of the CII repressor in the cryptic prophage CP-933P from Escherichia coli
O157:H7
Acta Crystallographica Section D Structural Biology 79(3):245-258 (2023)
Prolič-Kalinšek M, Volkov A, Hadži S, Van Dyck J, Bervoets I, Charlier D, Loris R
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
130 |
nm3 |
|
|
UniProt ID: A0A6M7H0F8 (2-141) YdaT_toxin domain-containing protein (mutant: L111N, F118R)
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDQC9_fit1_model1.png)
|
| Sample: |
YdaT_toxin domain-containing protein (mutant: L111N, F118R) monomer, 18 kDa Escherichia coli O157:H7 protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Apr 14
|
Structural basis of DNA binding by YdaT, a functional equivalent of the CII repressor in the cryptic prophage CP-933P from Escherichia coli
O157:H7
Acta Crystallographica Section D Structural Biology 79(3):245-258 (2023)
Prolič-Kalinšek M, Volkov A, Hadži S, Van Dyck J, Bervoets I, Charlier D, Loris R
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
UniProt ID: A0A6M7H0F8 (2-96) YdaT_toxin domain-containing protein
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDQD9_fit1_model1.png)
|
| Sample: |
YdaT_toxin domain-containing protein monomer, 13 kDa Escherichia coli O157:H7 protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Apr 16
|
Structural basis of DNA binding by YdaT, a functional equivalent of the CII repressor in the cryptic prophage CP-933P from Escherichia coli
O157:H7
Acta Crystallographica Section D Structural Biology 79(3):245-258 (2023)
Prolič-Kalinšek M, Volkov A, Hadži S, Van Dyck J, Bervoets I, Charlier D, Loris R
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.7 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
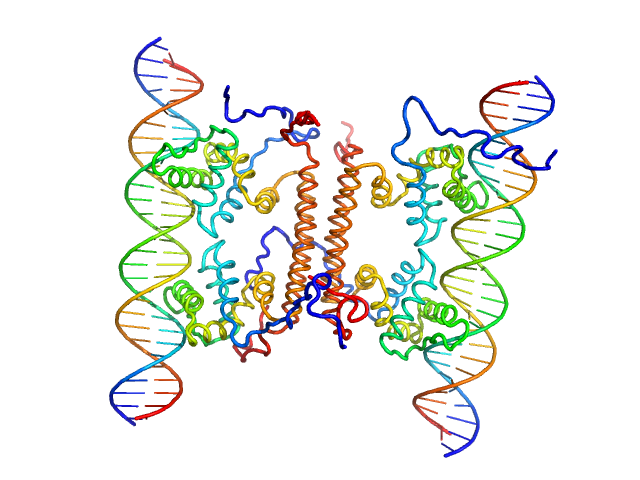
UniProt ID: A0A6M7H0F8 (2-141) YdaT_toxin domain-containing protein
UniProt ID: None (None-None) Om 30 base pair dsDNA
|
|
|
|
| Sample: |
YdaT_toxin domain-containing protein tetramer, 74 kDa Escherichia coli O157:H7 protein
Om 30 base pair dsDNA dimer, 37 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 14
|
Structural basis of DNA binding by YdaT, a functional equivalent of the CII repressor in the cryptic prophage CP-933P from Escherichia coli
O157:H7
Acta Crystallographica Section D Structural Biology 79(3):245-258 (2023)
Prolič-Kalinšek M, Volkov A, Hadži S, Van Dyck J, Bervoets I, Charlier D, Loris R
|
| RgGuinier |
4.2 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
190 |
nm3 |
|
|
UniProt ID: P08113 (22-754) Endoplasmin
|
|
|
|
| Sample: |
Endoplasmin dimer, 169 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 200 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Jul 11
|
Visualization of conformational transition of GRP94 in solution.
Life Sci Alliance 7(2) (2024)
Sun S, Zhu R, Zhu M, Wang Q, Li N, Yang B
|
| RgGuinier |
5.6 |
nm |
| Dmax |
24.0 |
nm |
| VolumePorod |
260 |
nm3 |
|
|
UniProt ID: P08113 (22-754) Endoplasmin
|
|
|
|
| Sample: |
Endoplasmin dimer, 169 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 200 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Jul 11
|
Visualization of conformational transition of GRP94 in solution.
Life Sci Alliance 7(2) (2024)
Sun S, Zhu R, Zhu M, Wang Q, Li N, Yang B
|
| RgGuinier |
5.7 |
nm |
| Dmax |
24.0 |
nm |
| VolumePorod |
274 |
nm3 |
|
|
UniProt ID: P08113 (22-754) Endoplasmin
|
|
|
|
| Sample: |
Endoplasmin dimer, 169 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 200 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Jul 11
|
Visualization of conformational transition of GRP94 in solution.
Life Sci Alliance 7(2) (2024)
Sun S, Zhu R, Zhu M, Wang Q, Li N, Yang B
|
| RgGuinier |
5.5 |
nm |
| Dmax |
24.5 |
nm |
| VolumePorod |
260 |
nm3 |
|
|
UniProt ID: P34710 (22-461) Netrin unc-6
UniProt ID: Q26261-1 (1-331) Isoform a of Netrin receptor unc-5
UniProt ID: None (None-None) Heparin, porcine intestinal mucosa
|
|
|
|
| Sample: |
Netrin unc-6 monomer, 52 kDa Caenorhabditis elegans protein
Isoform a of Netrin receptor unc-5 monomer, 38 kDa Caenorhabditis elegans protein
Heparin, porcine intestinal mucosa monomer, 15 kDa Sus scrofa domesticus
|
| Buffer: |
10 mM HEPES pH 7.2, 150 mM NaCl, 100 mM MgSO4, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Jun 18
|
Structural insights into the formation of repulsive netrin guidance complexes.
Sci Adv 10(7):eadj8083 (2024)
Priest JM, Nichols EL, Smock RG, Hopkins JB, Mendoza JL, Meijers R, Shen K, Özkan E
|
| RgGuinier |
10.4 |
nm |
| Dmax |
37.3 |
nm |
| VolumePorod |
2156 |
nm3 |
|
|
UniProt ID: P34710 (22-461) Netrin unc-6
UniProt ID: Q26261-1 (1-331) Isoform a of Netrin receptor unc-5
UniProt ID: None (None-None) Heparin, porcine intestinal mucosa
UniProt ID: G5EF96 (23-1082) Netrin receptor unc-40
|
|
|
|
| Sample: |
Netrin unc-6 monomer, 52 kDa Caenorhabditis elegans protein
Isoform a of Netrin receptor unc-5 monomer, 38 kDa Caenorhabditis elegans protein
Heparin, porcine intestinal mucosa monomer, 15 kDa Sus scrofa domesticus
Netrin receptor unc-40 monomer, 118 kDa Caenorhabditis elegans protein
|
| Buffer: |
10 mM HEPES pH 7.2, 150 mM NaCl, 100 mM MgSO4, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Dec 11
|
Structural insights into the formation of repulsive netrin guidance complexes.
Sci Adv 10(7):eadj8083 (2024)
Priest JM, Nichols EL, Smock RG, Hopkins JB, Mendoza JL, Meijers R, Shen K, Özkan E
|
| RgGuinier |
10.6 |
nm |
| Dmax |
36.5 |
nm |
| VolumePorod |
3410 |
nm3 |
|
|