UniProt ID: P42858 (1-88) Huntingtin
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDQS8_fit1_model1.png)
|
| Sample: |
Huntingtin monomer, 43 kDa Homo sapiens protein
|
| Buffer: |
20mM BisTris-HCl, 150mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Oct 28
|
The structure of pathogenic huntingtin exon 1 defines the bases of its aggregation propensity.
Nat Struct Mol Biol (2023)
Elena-Real CA, Sagar A, Urbanek A, Popovic M, Morató A, Estaña A, Fournet A, Doucet C, Lund XL, Shi ZD, Costa L, Thureau A, Allemand F, Swenson RE, Milhiet PE, Crehuet R, Barducci A, Cortés J, Sinnaeve D, Sibille N, Bernadó P
|
|
|
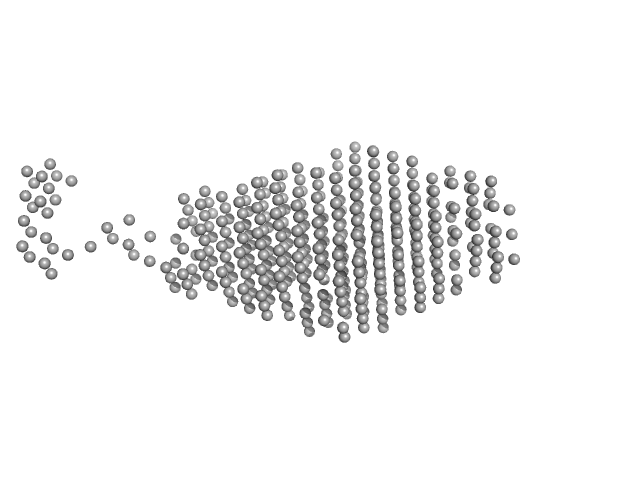
UniProt ID: A0A2S5LZS0 (3-314) Sensor domain-containing diguanylate cyclase
|
|
|
|
| Sample: |
Sensor domain-containing diguanylate cyclase dimer, 72 kDa Methylotenera sp. protein
|
| Buffer: |
10 mM Tris, 50 mM NaCl, 2 mM MgCl2, 3% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Apr 24
|
LOV-activated diguanylate cyclase
Ursula Vide
|
| RgGuinier |
3.4 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
128 |
nm3 |
|
|
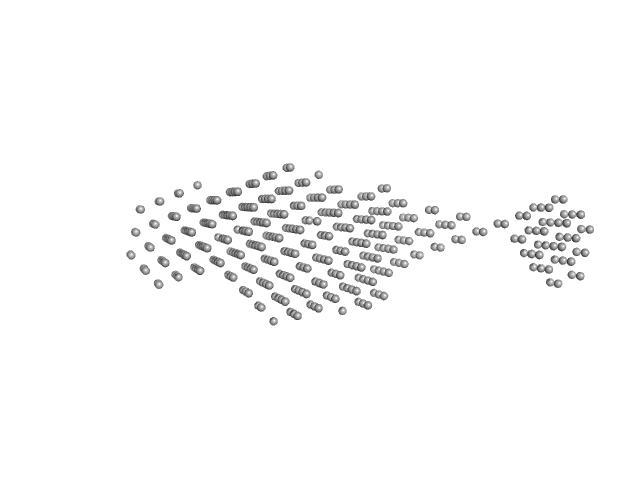
UniProt ID: A0A2S5LZS0 (3-314) Sensor domain-containing diguanylate cyclase
|
|
|
|
| Sample: |
Sensor domain-containing diguanylate cyclase dimer, 72 kDa Methylotenera sp. protein
|
| Buffer: |
10 mM Tris, 50 mM NaCl, 2 mM MgCl2, 3% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Apr 24
|
LOV-activated diguanylate cyclase
|
| RgGuinier |
5.2 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
159 |
nm3 |
|
|
UniProt ID: A0A0B4K6R7 (434-697) Na/Ca-exchange protein, isoform D
|
|
|
|
| Sample: |
Na/Ca-exchange protein, isoform D monomer, 31 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM TRIS, 200 mM NaCl, 1% v/v glycerol, 0.03% w/v NaN3, 1 mM β-mercaptoethanol, 2 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss, Institute of Physics, University of São Paulo on 2018 Mar 8
|
SAXS characterisation of CALX1.2 CBD12 construct
Roberto Kopke Salinas
|
| RgGuinier |
2.6 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
UniProt ID: A0A0B4K6R7 (434-697) Na/Ca-exchange protein, isoform D
|
|
|
|
| Sample: |
Na/Ca-exchange protein, isoform D monomer, 31 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM TRIS, 200 mM NaCl, 1% v/v glycerol, 0.03% w/v NaN3, 1 mM β-mercaptoethanol, 50 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss, Institute of Physics, University of São Paulo on 2018 Mar 8
|
SAXS characterisation of CALX1.2 CBD12 construct
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
UniProt ID: P02769 (25-607) Albumin
|
|
|
|
| Sample: |
Albumin monomer, 66 kDa Bos taurus protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SANS
data collected at SPB/SFX, European XFEL on 2021 May 14
|
Form factor determination of biological molecules with X-ray free electron laser small-angle scattering (XFEL-SAS).
Commun Biol 6(1):1057 (2023)
Blanchet CE, Round A, Mertens HDT, Ayyer K, Graewert M, Awel S, Franke D, Dörner K, Bajt S, Bean R, Custódio TF, de Wijn R, Juncheng E, Henkel A, Gruzinov A, Jeffries CM, Kim Y, Kirkwood H, Kloos M, Knoška J, Koliyadu J, Letrun R, Löw C, Makroczyova J, Mall A, Meijers R, Pena Murillo GE, Oberthür D, Round E, Seuring C, Sikorski M, Vagovic P, Valerio J, Wollweber T, Zhuang Y, Schulz J, Haas H, Chapman HN, Mancuso AP, Svergun D
|
|
|
UniProt ID: P02769 (25-607) Albumin
|
|
|
|
| Sample: |
Albumin monomer, 66 kDa Bos taurus protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SANS
data collected at SPB/SFX, European XFEL on 2021 May 15
|
Form factor determination of biological molecules with X-ray free electron laser small-angle scattering (XFEL-SAS).
Commun Biol 6(1):1057 (2023)
Blanchet CE, Round A, Mertens HDT, Ayyer K, Graewert M, Awel S, Franke D, Dörner K, Bajt S, Bean R, Custódio TF, de Wijn R, Juncheng E, Henkel A, Gruzinov A, Jeffries CM, Kim Y, Kirkwood H, Kloos M, Knoška J, Koliyadu J, Letrun R, Löw C, Makroczyova J, Mall A, Meijers R, Pena Murillo GE, Oberthür D, Round E, Seuring C, Sikorski M, Vagovic P, Valerio J, Wollweber T, Zhuang Y, Schulz J, Haas H, Chapman HN, Mancuso AP, Svergun D
|
|
|
UniProt ID: P02791 (1-175) Ferritin light chain
|
|
|
|
| Sample: |
Ferritin light chain 24-mer, 479 kDa Equus caballus protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SANS
data collected at SPB/SFX, European XFEL on 2021 May 14
|
Form factor determination of biological molecules with X-ray free electron laser small-angle scattering (XFEL-SAS).
Commun Biol 6(1):1057 (2023)
Blanchet CE, Round A, Mertens HDT, Ayyer K, Graewert M, Awel S, Franke D, Dörner K, Bajt S, Bean R, Custódio TF, de Wijn R, Juncheng E, Henkel A, Gruzinov A, Jeffries CM, Kim Y, Kirkwood H, Kloos M, Knoška J, Koliyadu J, Letrun R, Löw C, Makroczyova J, Mall A, Meijers R, Pena Murillo GE, Oberthür D, Round E, Seuring C, Sikorski M, Vagovic P, Valerio J, Wollweber T, Zhuang Y, Schulz J, Haas H, Chapman HN, Mancuso AP, Svergun D
|
|
|
UniProt ID: P01267 (1-2769) Thyroglobulin
|
|
|
|
| Sample: |
Thyroglobulin monomer, 303 kDa Bos taurus protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SANS
data collected at SPB/SFX, European XFEL on 2021 May 14
|
Form factor determination of biological molecules with X-ray free electron laser small-angle scattering (XFEL-SAS).
Commun Biol 6(1):1057 (2023)
Blanchet CE, Round A, Mertens HDT, Ayyer K, Graewert M, Awel S, Franke D, Dörner K, Bajt S, Bean R, Custódio TF, de Wijn R, Juncheng E, Henkel A, Gruzinov A, Jeffries CM, Kim Y, Kirkwood H, Kloos M, Knoška J, Koliyadu J, Letrun R, Löw C, Makroczyova J, Mall A, Meijers R, Pena Murillo GE, Oberthür D, Round E, Seuring C, Sikorski M, Vagovic P, Valerio J, Wollweber T, Zhuang Y, Schulz J, Haas H, Chapman HN, Mancuso AP, Svergun D
|
|
|
UniProt ID: P0DTC2 (319-566) Spike glycoprotein (ACE2 receptor binding domain)
|
|
|
|
| Sample: |
Spike glycoprotein (ACE2 receptor binding domain) monomer, 29 kDa Severe acute respiratory … protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SANS
data collected at SPB/SFX, European XFEL on 2021 May 14
|
Form factor determination of biological molecules with X-ray free electron laser small-angle scattering (XFEL-SAS).
Commun Biol 6(1):1057 (2023)
Blanchet CE, Round A, Mertens HDT, Ayyer K, Graewert M, Awel S, Franke D, Dörner K, Bajt S, Bean R, Custódio TF, de Wijn R, Juncheng E, Henkel A, Gruzinov A, Jeffries CM, Kim Y, Kirkwood H, Kloos M, Knoška J, Koliyadu J, Letrun R, Löw C, Makroczyova J, Mall A, Meijers R, Pena Murillo GE, Oberthür D, Round E, Seuring C, Sikorski M, Vagovic P, Valerio J, Wollweber T, Zhuang Y, Schulz J, Haas H, Chapman HN, Mancuso AP, Svergun D
|
|
|