UniProt ID: Q90922 (26-458) Netrin-1
UniProt ID: None (None-None) Heparin oligosaccharide dp10 ammonium salt
|
|
|
|
| Sample: |
Netrin-1 pentamer, 248 kDa Gallus gallus protein
Heparin oligosaccharide dp10 ammonium salt monomer, 3 kDa Sus scrofa domesticus
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Nov 26
|
The dynamic nature of netrin-1 and the structural basis for glycosaminoglycan fragment-induced filament formation
Nature Communications 14(1) (2023)
Meier M, Gupta M, Akgül S, McDougall M, Imhof T, Nikodemus D, Reuten R, Moya-Torres A, To V, Ferens F, Heide F, Padilla-Meier G, Kukura P, Huang W, Gerisch B, Mörgelin M, Poole K, Antebi A, Koch M, Stetefeld J
|
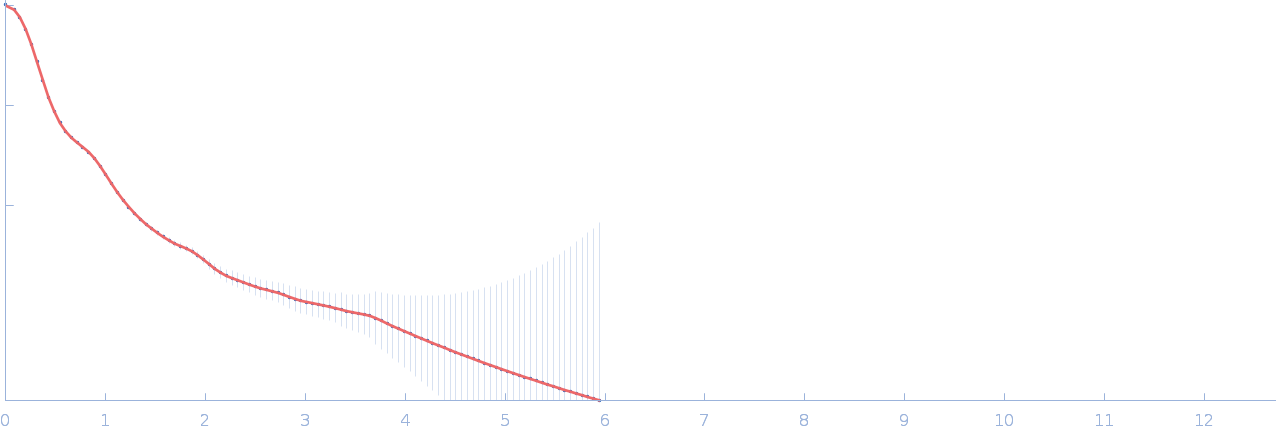
| RgGuinier |
6.5 |
nm |
| Dmax |
21.9 |
nm |
| VolumePorod |
769 |
nm3 |
|
|
UniProt ID: None (None-None) Ac-(POG)4-QG-(POG)5-NH2
|
|
|

|
| Sample: |
Ac-(POG)4-QG-(POG)5-NH2 trimer, 11 kDa synthetic construct protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, 2.7 mM KCL,, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Feb 1
|
A solution structure analysis reveals a bent collagen triple helix in the complement activation recognition molecule mannan-binding lectin.
J Biol Chem 299(2):102799 (2023)
Iqbal H, Fung KW, Gor J, Bishop AC, Makhatadze GI, Brodsky B, Perkins SJ
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
7 |
nm3 |
|
|
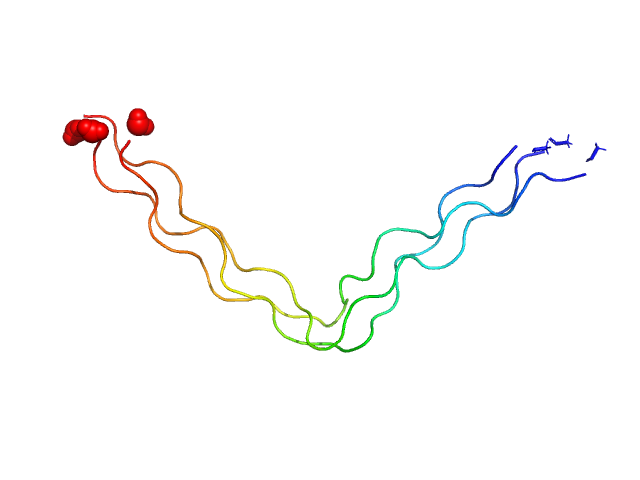
UniProt ID: None (None-None) Ac-(POG)4-QG-(POG)5-NH2
|
|
|
|
| Sample: |
Ac-(POG)4-QG-(POG)5-NH2 trimer, 11 kDa synthetic construct protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, 2.7 mM KCL,, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Feb 1
|
A solution structure analysis reveals a bent collagen triple helix in the complement activation recognition molecule mannan-binding lectin.
J Biol Chem 299(2):102799 (2023)
Iqbal H, Fung KW, Gor J, Bishop AC, Makhatadze GI, Brodsky B, Perkins SJ
|
| RgGuinier |
1.7 |
nm |
| Dmax |
7.1 |
nm |
| VolumePorod |
4 |
nm3 |
|
|
UniProt ID: None (None-None) Stem loop 2 and 3 in the 5'-genomic end of SARS-CoV-2
UniProt ID: P0DTC9 (44-180) Nucleoprotein
|
|
|
|
| Sample: |
Stem loop 2 and 3 in the 5'-genomic end of SARS-CoV-2 monomer, 14 kDa Severe acute respiratory … RNA
Nucleoprotein dimer, 30 kDa Severe acute respiratory … protein
|
| Buffer: |
25 mM HEPES, 75 mM KCl, 2.5 mM NaNO3, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Nov 29
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements
Karthikeyan Dhamotharan
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
UniProt ID: P0DTC9 (44-180) Nucleoprotein
UniProt ID: None (None-None) Stem loop 4 with AU extension in the 5'-genomic end of SARS-CoV-2
|
|
|
|
| Sample: |
Nucleoprotein monomer, 15 kDa Severe acute respiratory … protein
Stem loop 4 with AU extension in the 5'-genomic end of SARS-CoV-2 monomer, 22 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM HEPES, 75 mM KCl, 2.5 mM NaNO3, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Nov 29
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements
Karthikeyan Dhamotharan
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
UniProt ID: None (None-None) AU extension in the 5'-genomic end of SARS-CoV-2
UniProt ID: P0DTC9 (44-180) Nucleoprotein
|
|
|
|
| Sample: |
AU extension in the 5'-genomic end of SARS-CoV-2 monomer, 7 kDa Severe acute respiratory … RNA
Nucleoprotein dimer, 30 kDa Severe acute respiratory … protein
|
| Buffer: |
25 mM HEPES, 75 mM KCl, 2.5 mM NaNO3, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Nov 29
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements
Karthikeyan Dhamotharan
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
UniProt ID: P0DTC9 (44-180) Nucleoprotein
UniProt ID: None (None-None) Stem loop 4 in the 5'-genomic end of SARS-CoV-2
|
|
|
|
| Sample: |
Nucleoprotein monomer, 15 kDa Severe acute respiratory … protein
Stem loop 4 in the 5'-genomic end of SARS-CoV-2 monomer, 14 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM HEPES, 75 mM KCl, 2.5 mM NaNO3, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Nov 29
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements
Karthikeyan Dhamotharan
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
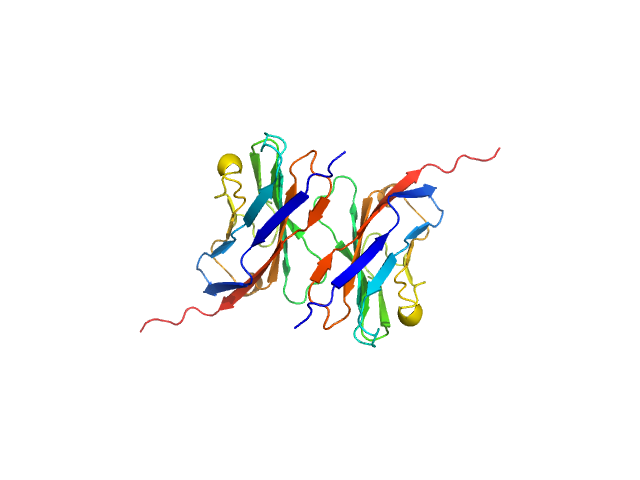
UniProt ID: P25189 (30-153) Myelin protein P0
|
|
|
|
| Sample: |
Myelin protein P0, 15 kDa Homo sapiens protein
|
| Buffer: |
50 mM NaCl, 1 mM EDTA, 20 mM TrisCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Jul 21
|
Homomeric interactions of the MPZ Ig domain and their relation to Charcot-Marie-Tooth disease
Brain (2023)
Ptak C, Peterson T, Hopkins J, Ahern C, Shy M, Piper R
|
| RgGuinier |
2.9 |
nm |
| Dmax |
17.0 |
nm |
|
|
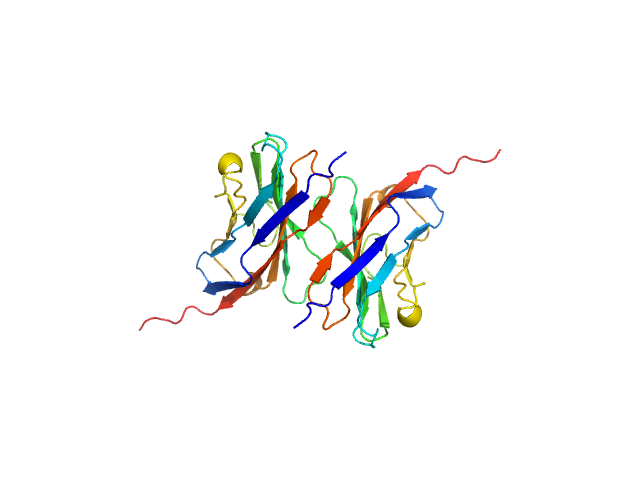
UniProt ID: P25189 (30-153) Myelin protein P0 (W53A, R74A, D75R)
|
|
|
|
| Sample: |
Myelin protein P0 (W53A, R74A, D75R), 14 kDa Homo sapiens protein
|
| Buffer: |
50 mM NaCl, 1 mM EDTA, 20 mM TrisCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Jul 14
|
Homomeric interactions of the MPZ Ig domain and their relation to Charcot-Marie-Tooth disease
Brain (2023)
Ptak C, Peterson T, Hopkins J, Ahern C, Shy M, Piper R
|
| RgGuinier |
1.9 |
nm |
| Dmax |
7.0 |
nm |
|
|
UniProt ID: P21980 (1-687) Protein-glutamine gamma-glutamyltransferase 2
|
|
|
|
| Sample: |
Protein-glutamine gamma-glutamyltransferase 2, 77 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 10% glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2021 Nov 19
|
Chaotic advection mixer for capturing transient states of diverse biological macromolecular systems with time-resolved small-angle X-ray scattering
IUCrJ 10(3):363-375 (2023)
Zielinski K, Katz A, Calvey G, Pabit S, Milano S, Aplin C, San Emeterio J, Cerione R, Pollack L
|
| RgGuinier |
4.1 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
150 |
nm3 |
|
|