UniProt ID: Q8XAD7 (1-100) Phage antirepressor protein Cro
|
|
|
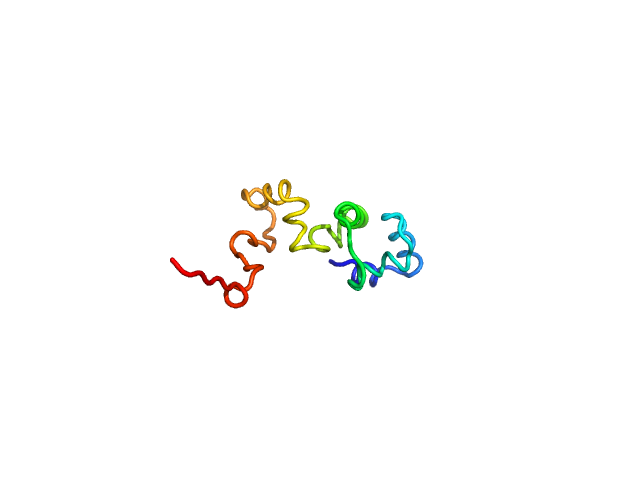
|
| Sample: |
Phage antirepressor protein Cro monomer, 12 kDa Escherichia coli O157:H7 protein
|
| Buffer: |
20 mM sodium acetate, 150 mM NaCl, 1 mM TCEP, pH: 5.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2018 Feb 5
|
Structural-function relationship of YdaS, a Cro-type repressor in the cryptic prophage CP-933P from Escherichia coli O157:H7
Marusa Prolic Kalinsek
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
19 |
nm3 |
|
|
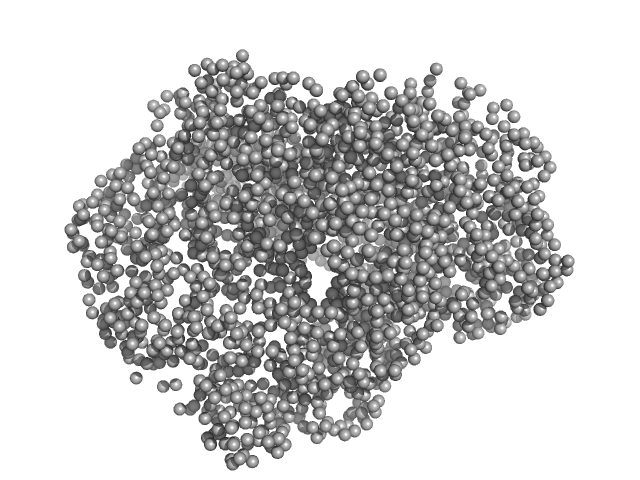
UniProt ID: K6TUQ9 (1-1038) Thiopeptide-type bacteriocin biosynthesis domain containing protein
|
|
|
|
| Sample: |
Thiopeptide-type bacteriocin biosynthesis domain containing protein dimer, 250 kDa Clostridium sp. Maddingley … protein
|
| Buffer: |
50 mM HEPES, 200 mM NaCl, 1 % (v/v) Glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 26
|
Structural insights into the substrate binding mechanism of the class I dehydratase MadB
Communications Biology 8(1) (2025)
Knospe C, Ortiz J, Reiners J, Kedrov A, Gertzen C, Smits S, Schmitt L
|
| RgGuinier |
4.5 |
nm |
| Dmax |
13.6 |
nm |
| VolumePorod |
440 |
nm3 |
|
|
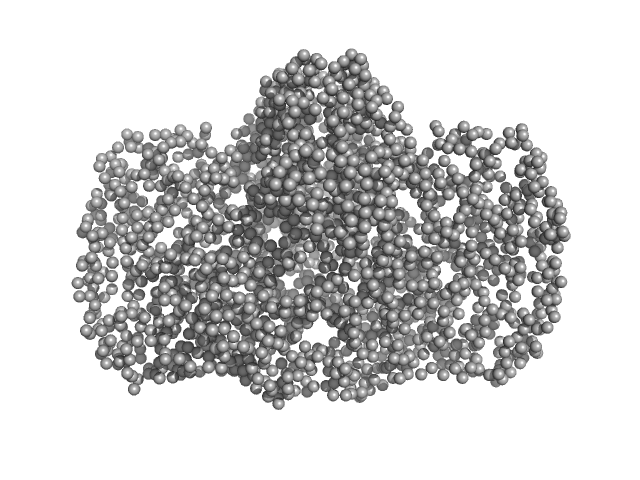
UniProt ID: K6TUQ9 (1-1038) Thiopeptide-type bacteriocin biosynthesis domain containing protein
UniProt ID: K6SWQ2 (1-33) Lantibiotic, gallidermin/nisin family
|
|
|
|
| Sample: |
Thiopeptide-type bacteriocin biosynthesis domain containing protein dimer, 250 kDa Clostridium sp. Maddingley … protein
Lantibiotic, gallidermin/nisin family monomer, 3 kDa Clostridium sp. Maddingley … protein
|
| Buffer: |
50 mM HEPES, 200 mM NaCl, 1 % (v/v) Glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Dec 3
|
Structural insights into the substrate binding mechanism of the class I dehydratase MadB
Communications Biology 8(1) (2025)
Knospe C, Ortiz J, Reiners J, Kedrov A, Gertzen C, Smits S, Schmitt L
|
| RgGuinier |
4.5 |
nm |
| Dmax |
13.8 |
nm |
| VolumePorod |
445 |
nm3 |
|
|
UniProt ID: None (1-76) HOTag-GS-Ubiquitin
|
|
|
|
| Sample: |
HOTag-GS-Ubiquitin tetramer, 51 kDa synthetic construct protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 16
|
Polyubiquitin ligand-induced phase transitions are optimized by spacing between ubiquitin units
Proceedings of the National Academy of Sciences 120(42) (2023)
Galagedera S, Dao T, Enos S, Chaudhuri A, Schmit J, Castañeda C
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
UniProt ID: None (1-76) HOTag6-(GS)2-Ubiquitin
|
|
|
|
| Sample: |
HOTag6-(GS)2-Ubiquitin tetramer, 52 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2023 Feb 21
|
Polyubiquitin ligand-induced phase transitions are optimized by spacing between ubiquitin units
Proceedings of the National Academy of Sciences 120(42) (2023)
Galagedera S, Dao T, Enos S, Chaudhuri A, Schmit J, Castañeda C
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.7 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
UniProt ID: None (1-76) HOTag-(GS)4-Ubiquitin
|
|
|
|
| Sample: |
HOTag-(GS)4-Ubiquitin tetramer, 53 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 16
|
Polyubiquitin ligand-induced phase transitions are optimized by spacing between ubiquitin units
Proceedings of the National Academy of Sciences 120(42) (2023)
Galagedera S, Dao T, Enos S, Chaudhuri A, Schmit J, Castañeda C
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
UniProt ID: None (1-76) HOTag-(GS)10-Ubiquitin
|
|
|
|
| Sample: |
HOTag-(GS)10-Ubiquitin tetramer, 56 kDa synthetic construct protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 16
|
Polyubiquitin ligand-induced phase transitions are optimized by spacing between ubiquitin units
Proceedings of the National Academy of Sciences 120(42) (2023)
Galagedera S, Dao T, Enos S, Chaudhuri A, Schmit J, Castañeda C
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.8 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
UniProt ID: None (1-76) HOTag6-(GS)25-Ubiquitin
|
|
|
|
| Sample: |
HOTag6-(GS)25-Ubiquitin tetramer, 65 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2023 Feb 21
|
Polyubiquitin ligand-induced phase transitions are optimized by spacing between ubiquitin units
Proceedings of the National Academy of Sciences 120(42) (2023)
Galagedera S, Dao T, Enos S, Chaudhuri A, Schmit J, Castañeda C
|
| RgGuinier |
4.7 |
nm |
| Dmax |
19.3 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
UniProt ID: None (1-76) HOTag-(GS)50-Ubiquitin
|
|
|
|
| Sample: |
HOTag-(GS)50-Ubiquitin tetramer, 80 kDa synthetic construct protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 16
|
Polyubiquitin ligand-induced phase transitions are optimized by spacing between ubiquitin units
Proceedings of the National Academy of Sciences 120(42) (2023)
Galagedera S, Dao T, Enos S, Chaudhuri A, Schmit J, Castañeda C
|
| RgGuinier |
5.6 |
nm |
| Dmax |
22.7 |
nm |
| VolumePorod |
193 |
nm3 |
|
|
UniProt ID: None (1-76) HOTag-PA-Ubiquitin
|
|
|
|
| Sample: |
HOTag-PA-Ubiquitin tetramer, 51 kDa synthetic construct protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 16
|
Polyubiquitin ligand-induced phase transitions are optimized by spacing between ubiquitin units
Proceedings of the National Academy of Sciences 120(42) (2023)
Galagedera S, Dao T, Enos S, Chaudhuri A, Schmit J, Castañeda C
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.9 |
nm |
| VolumePorod |
73 |
nm3 |
|
|